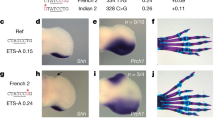
Abstract
Previously we have shown that nonsyndromic cleft lip with or without cleft palate (NSCL/P)1 is strongly associated with SNPs in IRF6 (interferon regulatory factor 6)2. Here, we use multispecies sequence comparisons to identify a common SNP (rs642961, G>A) in a newly identified IRF6 enhancer. The A allele is significantly overtransmitted (P = 1 × 10−11) in families with NSCL/P, in particular those with cleft lip but not cleft palate. Further, there is a dosage effect of the A allele, with a relative risk for cleft lip of 1.68 for the AG genotype and 2.40 for the AA genotype. EMSA and ChIP assays demonstrate that the risk allele disrupts the binding site of transcription factor AP-2α and expression analysis in the mouse localizes the enhancer activity to craniofacial and limb structures. Our findings place IRF6 and AP-2α in the same developmental pathway and identify a high-frequency variant in a regulatory element contributing substantially to a common, complex disorder.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Jugessur, A. & Murray, J.C. Orofacial clefting: recent insights into a complex trait. Curr. Opin. Genet. Dev. 15, 270–278 (2005).
Zucchero, T.M. et al. Interferon regulatory factor 6 (IRF6) gene variants and the risk of isolated cleft lip or palate. N. Engl. J. Med. 351, 769–780 (2004).
Kondo, S. et al. Mutations in IRF6 cause Van der Woude and popliteal pterygium syndromes. Nat. Genet. 32, 285–289 (2002).
Scapoli, L. et al. Strong evidence of linkage disequilibrium between polymorphisms at the IRF6 locus and nonsyndromic cleft lip with or without cleft palate, in an Italian population. Am. J. Hum. Genet. 76, 180–183 (2005).
Blanton, S.H. et al. Variation in IRF6 contributes to nonsyndromic cleft lip and palate. Am. J. Med. Genet. A. 137, 259–262 (2005).
Ghassibe, M. et al. Interferon regulatory factor-6: a gene predisposing to isolated cleft lip with or without cleft palate in the Belgian population. Eur. J. Hum. Genet. 13, 1239–1242 (2005).
Park, J.W. et al. Association between IRF6 and nonsyndromic cleft lip with or without cleft palate in four populations. Genet. Med. 9, 219–227 (2007).
Schorle, H., Meier, P., Buchert, M., Jaenisch, R. & Mitchell, P.J. Transcription factor AP-2 essential for cranial closure and craniofacial development. Nature 381, 235–238 (1996).
Milunsky, J.M. et al. TFAP2A mutations result in branchio-oculo-facial syndrome. Am. J. Hum. Genet. 82, 1171–1177 (2008).
Horvath, S., Xu, X. & Laird, N.M. The family based association test method: strategies for studying general genotype–phenotype associations. Eur. J. Hum. Genet. 9, 301–306 (2001).
Graham, R.R. et al. Three functional variants of IFN regulatory factor 5 (IRF5) define risk and protective haplotypes for human lupus. Proc. Natl. Acad. Sci. USA 104, 6758–6763 (2007).
Bille, C. et al. Oral clefts and life style factors–a case-cohort study based on prospective Danish data. Eur. J. Epidemiol. 22, 173–181 (2007).
Nguyen, R.H., Wilcox, A.J., Moen, B.E., McConnaughey, D.R. & Lie, R.T. Parent's occupation and isolated orofacial clefts in Norway: a population-based case-control study. Ann. Epidemiol. 17, 763–771 (2007).
Mitchell, L.E. et al. Guidelines for the design and analysis of studies on nonsyndromic cleft lip and cleft palate in humans: summary report from a Workshop of the International Consortium for Oral Clefts Genetics. Cleft Palate Craniofac. J. 39, 93–100 (2002).
Harville, E.W., Wilcox, A.J., Lie, R.T., Vindenes, H. & Abyholm, F. Cleft lip and palate versus cleft lip only: are they distinct defects? Am. J. Epidemiol. 162, 448–453 (2005).
Marazita, M.L. et al. Meta-analysis of 13 genome scans reveals multiple cleft lip/palate genes with novel loci on 9q21 and 2q32–35. Am. J. Hum. Genet. 75, 161–173 (2004).
Mossey, P. Epidemiology underpinning research in the aetiology of orofacial clefts. Orthod. Craniofac. Res. 10, 114–120 (2007).
Poulin, F. et al. In vivo characterization of a vertebrate ultraconserved enhancer. Genomics 85, 774–781 (2005).
Knight, A.S., Schutte, B.C., Jiang, R. & Dixon, M.J. Developmental expression analysis of the mouse and chick orthologues of IRF6: the gene mutated in Van der Woude syndrome. Dev. Dyn. 235, 1441–1447 (2006).
Sharpe, J. et al. Optical projection tomography as a tool for 3D microscopy and gene expression studies. Science 296, 541–545 (2002).
Cirulli, E.T. & Goldstein, D.B. In vitro assays fail to predict in vivo effects of regulatory polymorphisms. Hum. Mol. Genet. 16, 1931–1939 (2007).
Vieira, A.R. et al. Medical sequencing of candidate genes for nonsyndromic cleft lip and palate. PLoS Genet. 1, e64 (2005).
Riley, B.M. et al. Impaired FGF signaling contributes to cleft lip and palate. Proc. Natl. Acad. Sci. USA 104, 4512–4517 (2007).
Thomas, J.W. et al. Comparative analyses of multi-species sequences from targeted genomic regions. Nature 424, 788–793 (2003).
Schwartz, S. et al. MultiPipMaker and supporting tools: alignments and analysis of multiple genomic DNA sequences. Nucleic Acids Res. 31, 3518–3524 (2003).
Margulies, E.H., Blanchette, M., Haussler, D. & Green, E.D. Identification and characterization of multi-species conserved sequences. Genome Res. 13, 2507–2518 (2003).
Nickerson, D.A., Tobe, V.O. & Taylor, S.L. PolyPhred: automating the detection and genotyping of single nucleotide substitutions using fluorescence-based resequencing. Nucleic Acids Res. 25, 2745–2751 (1997).
Weinberg, C.R. Methods for detection of parent-of-origin effects in genetic studies of case-parents triads. Am. J. Hum. Genet. 65, 229–235 (1999).
Dudbridge, F. Likelihood-based association analysis for nuclear families and unrelated subjects with missing genotype data. Hum. Hered. 66, 87–98 (2008).
Provenzano, M.J. et al. AP-2 participates in the transcriptional control of the amyloid precursor protein (APP) gene in oral squamous cell carcinoma. Exp. Mol. Pathol. 83, 277–282 (2007).
Acknowledgements
We would like to thank A. Kinoshita, K. Frees, A. Mansilla, J. L'Heureux, M. Johnson, H. Morrison, G. Wehby, N. Rorick, K. Bedell and L. Powers for technical assistance and S. McConnell, D. Benton and M. DeVore for their administrative assistance. We would also like to thank A. Klingelhutz (University of Iowa) for kindly providing us with HFK cell line. This work was supported by grants from the National Institutes of Health (NIH): P50 DE16215 (J.C.M., M.L.M., B.C.S.), P30 ES05605 (J.C.M.), R37 DE08559 (J.C.M., M.L.M.), R01-DE13513 (B.C.S.), 1 UL1 RR024979-01 (J.C.M., B.C.S.), R01-CA73612 (F.E.D.), R01-HG003988 administered under Department of Energy Contract DE-AC02-05CH11231 (L.A.P.) as well as by the Intramural Research Program of the National Human Genome Research Institute (E.D.G.), in part by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences (A.J.W.), and European Commission FP5: EUROCRAN Project (contract no. QLG1-CT-2000-01019) (M.R., P.A.M., J.L., R.P.S.-T.). A.V. was supported by the American Heart Association. M.J.H. received salary support from T32 CA078586.
Author information
Authors and Affiliations
Consortia
Contributions
F.R. performed sequencing, SNP genotyping and statistical analysis, comparative sequence analysis, functional assays, drafted the manuscript and prepared figures and tables including supplementary material. M.L.M. conducted the statistical analyses, wrote several sections and edited the manuscript. M.E.C. and M.G. assisted with the statistical analyses. A.V., L.A.P. and D.R.F. performed transgenic mouse enhancer assay and contributed critical revisions. M.J.H. and F.E.D. provided samples for ChIP assay. E.D.G. and NISC generated sequences for comparative sequence analysis. M.R., K.C., C.B., M.M., A.J., R.T.L. A.J.W., P.A.M., J.L. and R.P.S.-T. coordinated the recruitment, phenotyping and sample collection. M.R. genotyped EUROCRAN samples. B.C.S. was involved in study design, data interpretation and contributed critical revisions. J.C.M. is the principal investigator and obtained funding, carried out patient recruitment, established key collaborations, provided study design and supervision and oversaw manuscript preparation and revision.
Corresponding author
Supplementary information
Supplementary Text and Figures
Supplementary Methods, Supplementary Tables 1–4, Supplementary Figures 1 and 2 (PDF 4608 kb)
Supplementary Video 1
Embryo 3a (MPG 27826 kb)
Supplementary Video 2
Embryo 3b (MPG 27838 kb)
Supplementary Video 3
Embryo 3c (MPG 20900 kb)
Rights and permissions
About this article
Cite this article
Rahimov, F., Marazita, M., Visel, A. et al. Disruption of an AP-2α binding site in an IRF6 enhancer is associated with cleft lip. Nat Genet 40, 1341–1347 (2008). https://doi.org/10.1038/ng.242
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.242
This article is cited by
-
Global perspectives in orofacial cleft management and research
British Dental Journal (2023)
-
Allele-specific transcription factor binding in a cellular model of orofacial clefting
Scientific Reports (2022)
-
Machine learning in prediction of genetic risk of nonsyndromic oral clefts in the Brazilian population
Clinical Oral Investigations (2021)
-
Systems genetics of nonsyndromic orofacial clefting provides insights into its complex aetiology
European Journal of Human Genetics (2019)
-
Systematic analysis of dark and camouflaged genes reveals disease-relevant genes hiding in plain sight
Genome Biology (2019)