Abstract
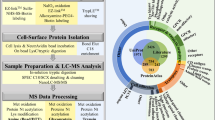
B-cell-specific plasma-membrane proteins are potential targets for either small molecule or antibody-based therapies. We have sought to annotate proteins expressed at the cell surface membrane in patients with chronic lymphocytic leukemia (CLL) using plasma-membrane-based proteomic analysis to identify previously uncharacterized and potentially B-cell-specific proteins. Proteins from plasma-membrane fractions were separated on one-dimensional gels and trypsinized fractions subjected to high-throughput MALDI-TOF mass spectrometry. Using this method, many known B-cell surface antigens were detected, but also known proteins not previously described in this disease or in this cellular compartment, including cell surface receptors, membrane-associated enzymes and secreted proteins, and completely unknown proteins. To validate the method, we show that BLK, a B-cell-specific kinase, is located in the CLL-plasma-membrane fraction. We also describe two novel proteins (MIG2B and B-cell novel protein #1, BCNP1), which are expressed preferentially in B cells. MIG2B is in a highly conserved and defined gene family containing two plasma-membrane-binding ezrin/radixin/moesin domains and a pleckstrin homology domain; the Caenorhabditis elegans homolog (UNC-112) is a membrane-associated protein that colocalizes with integrin at cell–matrix adhesion complexes. BCNP1 is a completely unknown protein with three predicted transmembrane domains, with three alternatively spliced final exons. Proteomic analysis may thus define new potential therapeutic targets.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Rozman C, Montserrat E . Current concepts: chronic lymphocytic leukemia. N Engl J Med 1995; 333: 1052–1057.
Kalil N, Cheson BD . Chronic lymphocytic leukemia. Oncologist 1999; 4: 352–369.
Aalto Y, El-Rifai W, Vilpo L, Ollila J, Nagy B, Vihinen M et al. Distinct gene expression profiling in chronic lymphocytic leukemia with 11q23 deletion. Leukemia 2001; 15: 1721–1728.
Stratowa C, Loffler G, Lichter P, Stilgenbauer S, Haberl P, Schweifer N et al. CDNA microarray gene expression analysis of B-cell chronic lymphocytic leukemia proposes potential new prognostic markers involved in lymphocyte trafficking. Int J Cancer 2001; 91: 474–480.
Korz C, Pscherer A, Benner A, Mertens D, Schaffner C, Leupolt E et al. Evidence for distinct pathomechanisms in B-cell chronic lymphocytic leukemia and mantle cell lymphoma by quantitative expression analysis of cell cycle and apoptosis-associated genes. Blood 2002; 99: 4554–4561.
Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature 2000; 403: 503–511.
Voss T, Ahorn H, Harbel P, Wilgenbus K . Correlation of clinical data with proteomics profiles in 24 patients with B-cell chronic lymphocytic leukemia. Int J Cancer 2000; 91: 180–186.
Pasquali C, Fialka I, Huber L-A . Subcellular fractionation, electromigration analysis and mapping of organelles. J Chromatogr B Biomed Sci Appl 1999; 722: 89–102.
Eng J-K, McCormack A-L, Yates III J-R . An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein. J Am Soc Mass Spectrom 1994; 5: 976–989.
Boyd RS, Duggan MJ, Shone CC, Foster KA . The effect of botulinum neurotoxins on the release of insulin from the insulinoma cell lines HIT-15 and RINm5F. J Biol Chem 1995; 270: 18216–18218.
Pappin DJC, Hojrup P, Bleasby AJ . Rapid identification of proteins by peptide mass fingerprinting. Curr Biol 1993; 3: 327–332, also http://www.hgmp.mrc.ac.uk/Bioinformatics/Webapp/mowse.
Adam PJ, Boyd R, Tyson KL, Fletcher GC, Stamps A, Hudson L et al. Comprehensive proteomic analysis of breast cancer cell membranes reveals unique proteins with potential roles in clinical cancer. J Biol Chem 2003; 278: 6482–6489.
Belov L, de la Vega O, dos Remedios CG, Mulligan SP, Christopherson RI . Immunotyping of leukemias using a cluster of differentiation antibody microarray. Cancer Res 2001; 61: 4483–4489.
Hulkkonen J, Vilpo L, Hurme M, Vilpo J . Surface antigen expression in chronic lymphocytic leukemia: clustering analysis, interrelationships and effects of chromosomal abnormalities. Leukemia 2002; 16: 178–185.
Treumann A, Lifely MR, Schneider P, Ferguson MA . Primary structure of CD52. J Biol Chem 1995; 270: 6088–6099.
Burge C, Karlin S . Prediction of complete gene structures in human genomic DNA. J Mol Biol 1997; 268: 78–94.
Venkataraman C, Muthusamy N, Muthukkumar S, Bondada S . Activation of lyn, blk, and btk but not syk in CD72-stimulated B lymphocytes. J Immunol 1998; 160: 3322–3329.
Majima S, Kajino K, Fukuda T, Otsuka F, Hino O . A novel gene ‘Niban’ upregulated in renal carcinogenesis cloning by the cDNA-amplified fragment length polymorphis approach. Jpn J Cancer Res 2000; 91: 869–874.
Wick M, Burger C, Brusselbach S, Lucibello FC, Muller R . Identification of serum-inducible genes: different patterns of gene regulation during G0 → S and G1 → S progression. J Cell Sci 1994; 107: 227–239.
Rogalski TM, Mullen GP, Gilbert MM, Williams BD, Moerman DG . The UNC-112 gene in Caenorhabditis elegans encodes a novel component of the cell–matrix adhesion structures required for integrin localization in the muscle membrane. J Cell Biol 2000; 150: 253–264.
Mackinnon AC, Qadota H, Norman KR, Moerman DG, Williams BD . C. elegans PAT-4/ILK functions as an adaptor protein within integrin adhesion complexes. Curr Biol 2002; 14: 787–797.
Acknowledgements
This work was supported in part by grants from the Kay Kendall Leukemia Fund and the Leukemia Research Fund.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Boyd, R., Adam, P., Patel, S. et al. Proteomic analysis of the cell-surface membrane in chronic lymphocytic leukemia: identification of two novel proteins, BCNP1 and MIG2B. Leukemia 17, 1605–1612 (2003). https://doi.org/10.1038/sj.leu.2402993
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.leu.2402993
Keywords
This article is cited by
-
Lck is a relevant target in chronic lymphocytic leukaemia cells whose expression variance is unrelated to disease outcome
Scientific Reports (2017)
-
Gene expression profiling of peripheral blood mononuclear cells in the setting of peripheral arterial disease
Journal of Clinical Bioinformatics (2012)
-
Correlation analysis of two-dimensional gel electrophoretic protein patterns and biological variables
BMC Bioinformatics (2006)
-
Proteomics in pathology research
Laboratory Investigation (2004)