Abstract
Objective: To investigate the incidence of mixed-species (MS) malaria infection, and compare the results with microscopically confirmed cases of malaria.
Methods: During 2010, blood spots collected from 371 clinically suspected cases of malaria were microscopically examined in a cross-sectional study. The DNA was extracted from the samples, and a nested polymerase chain reaction (PCR) was performed. The results obtained by the 2 methods were compared.
Results: From the microscopic analysis it was determined that 369 samples (99.5%) were positive for Plasmodium falciparum (P. falciparum) and 2 were Plasmodium vivax (P. vivax) mono-infections. There were no mixed malaria infections. The PCR analysis, however, showed that in 7 cases (1.9%) the infection was caused by MS malaria comprising of P. falciparum and P. vivax, 2 of these representing the cases that were microscopically diagnosed as P. vivax mono-infections. All cases were negative for Plasmodium malariae, Plasmodium ovale, and Plasmodium knowlesi.
Conclusion: Mixed malaria infections are currently overlooked when using microscopy. The PCR assays are essential complementary techniques that should be used with microscopic examination of blood smears.
Malaria is among the most prevalent of endemic tropical diseases. Human infection with malaria is caused by 4 species of plasmodia: Plasmodium falciparum (P. falciparum), Plasmodium vivax (P. vivax), Plasmodium malariae (P. malariae), Plasmodium ovale (P. ovale), and, rarely, Plasmodium knowlesi (P. knowlesi). Each of these species is associated with different clinical features and outcomes. The risk of severe malaria symptoms increases considerably if treatment is delayed. In 2012, 99 countries documented ongoing malaria transmission with around 3.3 billion people at risk of contracting the disease. Globally, 219 million cases of malaria were reported with 660,000 associated deaths.1 In Africa, the predominant malaria species is P. falciparum. Elsewhere; however, including Russia, the tropical regions of Asia, the Pacific, and South, and Central America, P. vivax is the most common species. In geographical areas where more than one malaria species is present, these infections may combine and such combined infections are usually under-reported.2 In the Kingdom of Saudi Arabia (KSA), Jazan and Aseer provinces, in the south-west of the country, are the most malaria-endemic areas with a prominence of P. falciparum. No mixed-infection cases have been reported from these regions.3 In early 2008, Artemisinin-based combination therapy (ACT) was introduced to KSA to treat all malaria-positive cases. For P. vivax malaria, chloroquine is used, but when P. vivax is resistant to chloroquine, an appropriate ACT regimen is recommended together with a primaquine regimen.4 Severe and complicated malaria is most commonly caused by P. falciparum, although P. vivax, and P. knowlesi may also be responsible for severe infections. Research on the P. vivax parasite has tended to lag behind, since it was previously thought that mono-infection with P. vivax resulted only in benign tertian fever, and that severe cases of P. vivax infection were the result of co-infection with P. falciparum. Recent evidence, however, suggests that clinical symptoms for this infection have changed and that P. vivax mono-infections can result in severe malaria (SM) and even death.5 Incorrect diagnosis of the malaria species causing infection in a patient is a potentially significant problem, since misdiagnosis could result in the infection becoming severe, in the case of P. falciparum, or relapse in the case of P. vivax.6 False-negative diagnoses for P. vivax are common in endemic areas, and many untreated patients therefore serve as reservoir hosts of malaria parasites.7 In addition, if P. vivax schizonts are detected in venous blood, co-infection with P. falciparum may be missed, since P. falciparum schizonts are only present in the capillaries of internal organs.7
Microscopic examination of Giemsa-stained blood films is the best technique for detecting the malaria parasite due to its low cost and ability to distinguish between malaria species. Nevertheless, accurate diagnosis depends on an experienced technician. Misdiagnosis can easily occur, particularly in cases involving mixed malarial infections or when parasitemia is low.8 To overcome these limitations, several diagnostic methods have been developed, including rapid diagnostic tests (RDT), serologic tests and direct polymerase chain reaction (PCR).8 Overall microscopy and RDTs comprise the main means of diagnosing the presence of malaria. When it is necessary to distinguish between the infecting species, PCR would be preferred. Of these approaches, PCR assays are most commonly used to detect the 5 species of plasmodia precisely. The PCR techniques involve a universal plasmodium primer based on the sequence of the small-subunit ribosomal RNA (ssrRNA) gene.9
The purpose of this study was to explore the prevalence of mixed-species infections of P. falciparum and P. vivax parasites as well as P. malariae, P. ovale, and P. knowlesi in Jazan Province - south-west Saudi Arabia by PCR and compare the results obtained with microscopically confirmed cases of malaria.
Methods
Sampling and malaria microscopy examination
In this cross-sectional study, 371 cases of clinically suspected malaria were studied in this research (n=371), all taken from patients referred to the Malaria Research Center, Jazan Province, KSA during 2010. Written consent was obtained from all study participants, and the study protocol was approved by King Khalid University, Abha, KSA. Thick and thin blood films were prepared and 3 drops of each patient’s blood were blotted onto a 3-mm Whatman® filter paper. The filter papers were carefully air-dried at room temperature and stored under sealed conditions at 4°C for additional processing
Molecular identification of malaria parasites and assessment of mixed malaria species
Genomic DNA was extracted from the blood spots collected on filter papers from all cases (n=371) using a Qiagen kit. Next, a filter paper disc was punched out of each blood spot and placed in a 1.5-ml centrifuge tube. The DNA was eluted using 50 µl of AE elution buffer and kept at -20°C until used in PCR assays. Plasmodium species were also identified by 18S rRNA-based nested PCR using genus- and species-specific primers as previously described.10 The products were analysed by gel electrophoresis using a 1.5% (weight in volume) agarose gel.
Statistical analysis
Sensitivity, specificity, and concomitant 95% confidence intervals were computed for microscopy in comparison with the molecular identification. To compensate for zero frequency, 0.5 was added to all cells in the contingency table.11 Statistical analysis was performed with the Statistical Package for Social Sciences (SPSS, version 10.0, Chicago, IL, USA).
Results
Microscopically, all specimens were positive for P. falciparum, except 2 that were diagnosed as P. vivax. When screened by molecular techniques using 18S rRNA-based nested PCR, 371 (100%) tested positive for P. falciparum (Figure 1), and 7 of them (1.9%) also tested positive for P. vivax (Figure 2), meaning that each case of P. vivax has P. falciparum parasites co-existing. This result contrasts with the health statistical report in Saudi Arabia in 2010,3 which showed that there were no cases of mixed malaria infection. Additionally, multiplex PCR assays proved that all samples were negative for P. malariae, P. ovale, and P. knowlesi.
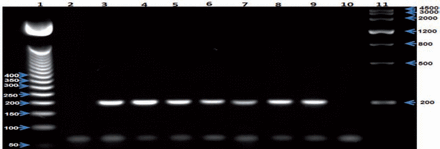
Second round of nested PCR to detect Plasmodium falciparum, indicating positivity at the expected size (205 bp). Lane 1- 50-bp DNA Step Ladder (Promega), lane 2 - negative control, lane 3 - positive control, lanes 4-9 - samples, lane 10 - control reagent; lane 11 - Gel Pilot Wide Range Ladder (100) (Qiagen).
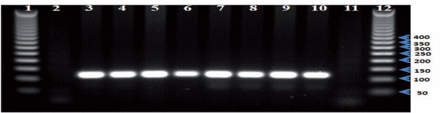
Second round of nested PCR to detection Plasmodium= vivax, indicating positivity at the expected size (120 bp). Lanes 1, 12 - 50-bp DNA Step Ladder (Promega), lane 2 - negative control, lane 3 - positive control, lanes 4–10 - samples, lane 11- control reagent.
Table 1 shows that the sensitivity of microscopy (the test’s ability to identify a condition correctly) in comparison with molecular testing was 99.9% for P. falciparum, but this decreased to just 31.25% for P. vivax. For mixed infections, the sensitivity dropped drastically to reach 6.25%. On the other hand, the specificity (namely the test’s ability to exclude a condition correctly) of microscopy in comparison with molecular testing was 50% for P. falciparum, increasing to reach 99.9% for P. vivax.
Sensitivity, specificity and concomitant 95% confidence intervals of microscopy in comparison to the molecular identification in screening for single or combined Plasmodium (P) falciparum and P. vivax.
Discussion
Although the incidence of malaria in KSA has decreased over the last decade, vector resistance to insecticides, drug resistance, and imported malaria can each lead to malaria transmission particularly with respect to P. falciparum and P. vivax infections. In addition, the Hajj and Umrah religious rituals may introduce malaria into KSA, as many Muslim countries have a high prevalence of P. falciparum and P. vivax malaria.4
Understanding these epidemiological patterns and species distribution dynamics is important for KSA, given that it is located between different eco-epidemiological malarial zones (Afrotropical, Oriental, and Palearctic). Imported cases from Southeast Asia (including India) as well as the Eastern Mediterranean region (including Djibouti, Sudan, and Yemen) may result in an influx of P. falciparum. However, most cases in Afghanistan, Pakistan, Iran, India, and Iraq result from infection by P. vivax. In the Western Pacific region, P. falciparum and P. vivax can be found, but malaria transmission is caused entirely by P. vivax in the Republic of Korea and in central areas of China.1
In this study, PCR analysis revealed 7 cases of mixed infection in Jazan that had not been detected by microscopy. Instead, microscopy indicated only 2 mono-infections with P. vivax, and no mixed infections in any province of KSA.3 This inconsistency suggests that many cases of mixed infections may be misdiagnosed where only microscopy-based diagnostic techniques are used. The findings of this study contradict several studies carried out in KSA that show P. falciparum infection to be more prevalent than other species and no mixed infections were reported, Dawoud et al’s study12 is an example of these studies.
In mixed falciparum/vivax infections, there is a tendency for one species to dominate. These mixed malaria infections are occasionally misdiagnosed with microscopic detection techniques, so there is a case for the use of more accurate diagnosis techniques, such as PCR.6 The inability to detect mixed infection with P. falciparum and P. vivax using microscopy may result from competition at the level of host red blood cells or cross-species immunity.6
Several studies have shown that PCR is more reliable than microscopy for the detection of malaria in areas of low parasitemia. This method is reproducible and can be completed with as few as 5 parasites per microlitre of blood. Thus, PCR is the ideal method for identifying mixed infections that would be overlooked using classical diagnostic methods.13 This study demonstrates that MS malaria was present in 1.7% of microscopically-confirmed cases with a single species infection. However, studies in Thailand demonstrated that 30% of patients with P. falciparum malaria suffered from symptomatic P. vivax malaria.14 A study in India showed 45.5% mixed P. falciparum/P. vivax by PCR assay,15 while another study in Iran showed that 14.8% of cases were mixed P. falciparum/P. vivax.16 Accurate diagnosis of malaria species is essential for both correct and adequate treatment and also for the design of an effective vaccine and other malaria control measures. Limitations of this study include a lack of comparison with RDT assays and the fact that it was conducted only in the South-West Provinces of Saudi Arabia.
In conclusion, little is known about MS malaria profiles in the Jazan Region of KSA. The use of PCR diagnosis, together with routine methods, will facilitate early and accurate diagnosis of malaria species. Establishment of a reference PCR lab to detect malaria species and to deal with questionable cases of malaria post-screening by microscopy is recommended. Further studies are needed to compare PCR with RDT when diagnosing mixed malaria infections.
Acknowledgment
I wish to thank patients from the malaria-endemic Jazan region who kindly contributed to this study. I am also grateful to Professors Mohammed N. Al-Ahdal and Ahmed A. Al-Qahtani, Department of Infection and Immunity, King Faisal Specialist Hospital and Research Center, Riyadh, Saudi Arabia for assistance with the molecular typing of parasites and Prof. Ahmed A. Mahfouz, College of Medicine, King Khalid University, Abha for assistance with statistical analysis.
Footnotes
Disclosure. The experimental work was partially funded by King Khalid University Scientific Deanship, Abha, Kingdom of Saudi Arabia. The author declares that there was no conflict of interest and this work was not presented in a conference proceeding.
- Received October 6, 2014.
- Accepted November 10, 2014.
- Copyright: © Saudi Medical Journal
This is an open-access article distributed under the terms of the Creative Commons Attribution-Noncommercial-Share Alike 3.0 Unported, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.