Abstract
Objectives: To assess the association between interleukin (IL)-10 -1082, -819, -592 polymorphisms and tuberculosis (TB) risk.
Methods: This study was conducted between July and October 2014 in West China Hospital, Chengdu, Sichuan, China. We searched and collected data from PUBMED, EMBASE, Web of Science, China National Knowledge Infrastructure, VIP, and WANGFANG up to October 2014.
Results: A total of 37 studies were enrolled, including 8625 TB cases, and 9928 healthy controls. The IL-10-1082G/A polymorphism was found to be associated with TB susceptibility in Caucasian (GG versus GA+AA, odds ratio [OR] - 1.83, 95% confidence interval [CI] - 1.03-3.24). The IL-10-819C/T polymorphism was related to TB susceptibility among Asians (C versus T, OR - 0.88, 95% CI - 0.81-0.97; CC versus TT: OR - 0.79, 95% CI - 0.64-0.97; CC+CT versus TT: OR - 0.87, 95% CI - 0.77-0.98; CC versus CT+TT: OR - 0.82, 95% CI - 0.68-0.98). The IL-10-592C/A polymorphism was in association with TB susceptibility in Asians (C versus A: OR - 0.74, 95% CI - 0.65-0.85; CC versus AA: OR - 0.55, 95% CI - 0.41-0.75; CA versus AA: OR - 0.73, 95% CI - 0.60-0.89; CC+CA versus AA: OR - 0.69, 95% CI 0.58-0.83; CA versus AA: OR - 0.66, 95% CI 0.51-0.86), Caucasian (C versus A: OR - 1.25, 95% CI - 1.08-1.45; CC versus CA+AA: OR-1.48, 95% CI - 1.16-1.89), and Europeans (C versus A: OR - 1.31, 95% CI - 1.02-1.67; CC versus AA: OR - 1.88, 95% CI - 1.05-3.37).
Conclusion: This meta-analysis suggests that IL-10-1082G/A, IL-819C/T, and IL-592C/A polymorphisms might be associated with TB susceptibility in certain ethnicities.
Tuberculosis (TB), caused by Mycobacterium tuberculosis (MT) has long plagued human beings, ranking as the second major cause of death from a single infectious agent following human immunodeficiency virus (HIV).1 According to the World Health Organization in 2012,1 there were approximately 8.6 million new TB cases, and 1.3 million deaths with the greatest burden of disease falling in developing countries. Nearly one-third of the world’s population is infected with MT, but only a small fraction (5-15%) of the population develop clinical disease during their lifetime.2 This clinical diversity on disease’ susceptibility suggests both genetic predisposition, and environmental factors may contribute to the risk of infection by this pathogen following exposure and progression to TB disease.3 Cytokines involved in the shifting of the T-helper (Th)1/Th2 balance are known to play crucial roles in the progression of MT infection.4 Interleukin (IL)-10, as an immunoregulatory cytokine functions by down-regulating the Th1 driven pro-inflammatory response.5 It can be utilized by certain intracellular pathogens, including MT, to debilitate the host immune response, which then leads to the failure of subsequent clearance from the host.6 Studies7 carried out in vivo reported that IL-10-deficient mice infected with MT exhibits enhanced anti-mycobacterial immunity, while over-expression of IL-10 reactivates chronic pulmonary TB, and reduces MT-induced apoptosis of murine macrophages.8-11 The relationship between IL-10 and TB was also proven by several research conducted in humans.12,13 For patients, especially those with anergic TB and have increased IL-10 production, it is reported that high level IL-10 plasma level was associated with shorter survival of TB patients.12,13 Up until now, several studies have identified the association of IL-10 gene polymorphisms with altered IL-10 production, especially in 3 single nucleotide polymorphisms (SNPs) at positions -1082, -819, and -592 from the transcription start site.14,15 Investigating the association of the IL-10 gene polymorphisms with TB susceptibility may promote our understanding of the pathogenesis of TB, and explain individual differences in TB risk. What more, it is worth mentioning that Tobin et al16 recently reported their success in optimizing the inflammatory response to mycobacterial infections through the host genotype-specific therapies, which may also add our attention to studies regarding the TB gene polymorphisms. Unfortunately, although in recent years, a number of molecular epidemiological studies have been conducted to evaluate the risk of TB associated with the polymorphisms of IL-10 gene, their findings are conflicting. Inconsistency of those results can be attributed to a small sample size, insufficient power, and ethnic diversity of the population. Hence, we undertook this meta-analysis to evaluate the association of IL-10 gene polymorphisms and TB risk. This report follows the PRISMA (Preferred Reporting Items For Systematic Reviews and Meta-Analyses) statement.17
Methods
Search methods
This study was conducted between July and October 2014 in West China Hospital, Chengdu, China. The following databases were searched without language restriction: PUBMED, EMBASE, Web of Science, VIP, China National Knowledge Infrastructure, and WANGFANG. The search strategies were as follows: ‘TB’ or ‘tuberculosis’ in combination with ‘polymorphism’ or ‘variant’ or ‘genotype’ or ‘allele’ or ‘mutation’, and in combination with”interleukin-10,” or “IL-10”. Furthermore, the reference lists of reports identified by this search strategy were also searched to select relevant articles. The last search was conducted in October 2014. The inclusion criteria were as follows: 1) case-control study; 2) genotype distribution in both cases and controls were available; and 3) evaluation of IL-10- 1082, -819, -592 polymorphisms and TB risk. Exclusion criteria were: 1) duplicate of previous publications; and 2) study design based on family, or sibling pairs.
Data extraction
Two investigators independently screened records initially identified by the title or abstract, and then independently reviewed these records for eligibility. Discrepancies were resolved by discussion and simultaneous reference to the relevant literature. The following variables were collected from studies: title, first author’s name, year and source of publication, country of origin, ethnic descent of the study population, numbers of eligible cases and controls, and genotype distributions in cases and controls, definition of cases, HIV status, and genotyping method. If data concerning the genotype distributions were not displayed in the article, the investigators would contact the primary author in an attempt to obtain the missing data.
Statistical analysis
Hardy-Weinberg Equilibrium (HWE) was tested in the controls via the Chi-square test, and it was considered statistically significant when p<0.05. Heterogeneity was assessed by the I2 based Q statistics, and I2 test. Fixed effect model was adopted if the result of the heterogeneity test was p>0.10. Otherwise the random effect model was used.18,19 Odds ratio (OR) and 95% confidence interval (CI) were used to assess the strength of association between IL-10 polymorphisms, and the risk of TB under allelic model, homozygote comparison, heterozygote comparison, dominant, and recessive genetic model. Stratified analyses were performed by ethnicity. Sensitivity analyses were also performed to assess the stability of the results by excluding studies deviating from HWE. Begg’s and Egger’s tests were used to test the possible publication bias each time, when more than 5 studies were pooled.20 All statistical analyses were carried out using STATA version 12.0 (Stata Corp, College Station, Texas, USA) using 2-sided p values.
Results
Study characteristics
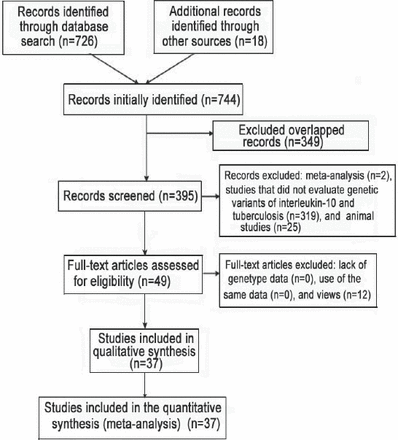
A total of 744 articles were initially identified. After excluding those that overlapped between the databases, 395 abstracts were retrieved for further evaluation. Then, 49 articles with full texts that met the inclusion criteria were assessed. Twelve articles were excluded for being reviews. Finally, a total of 37 studies, 31 in English, 5 in Chinese, and 1 in Turkish were included in this meta-analysis.21-57 Figure 1 provides a PRISMA flow diagram of the search. The characteristics of these 37 included studies are summarized in Table 1. They included 8625 cases and 9928 controls. Among them, there were 33 studies for the -1082G/A, 21 studies for the -819C/T, and 18 studies for the -592C/A. All the studies selected healthy people as controls. The study participants were diverse in ethnicity including African, Asian, European and Caucasian. The genotype distributions among the controls of all studies were consistent with HWE except for 11 studies for the -1082G/A, 4 studies for the -819C/T, and 4 studies for the -592C/A.
Flow diagram of studies included in a meta-analysis from China.
Characteristics of literature included in the meta-analysis on interleukin polymorphisms and tuberculosis risk study from China.
Quantitative synthesis of IL-10-1082G/A
The evaluation of association between IL-10-1082G/A polymorphism and TB risk are listed in Table 2. Overall, the results of pooling all studies showed no significant associations between the IL-10-1082 G/A polymorphism and TB risk under any of 5 genetic models (G versus A: OR - 1.04, 95% CI - 0.93-1.17; GG versus AA: OR - 1.27, 95% CI - 0.96-1.68; GA versus AA: OR - 1.02, 95% CI - 0.86-1.21; GG+GA versus AA: OR - 1.05, 95% CI - 0.88-1.25; GG versus GA+AA: OR - 1.22, 95% CI - 0.96-1.56). Excluding studies deviating from HWE did not change the results (G versus A: OR - 0.97, 95% CI - 0.84-1.13; GG versus AA: OR - 1.08, 95% CI - 0.75-1.55; GA versus AA: OR - 0.94, 95% CI - 0.78-1.14; GG+GA versus AA: OR - 0.97, 95% CI - 0.80-1.17; GG versus GA+AA: OR - 1.05, 95% CI - 0.77-1.44). We further performed subgroup analyses in IL-10-1082 G/A polymorphism by ethnicity, and no association of IL-10-1082G/A polymorphism and TB risk was observed in Asian and African descendants. While statistically significant associations were found in European descendants under 3 genetic models (G versus A: OR - 1.38, 95% CI - 1.04-1.82; GG versus AA: OR - 2.08, 95% CI 1.02-4.25; GG versus GA+AA: OR - 1.80, 95% CI - 1.28-2.54), and in Caucasian descendants under one genetic models (GG versus GA+AA: OR - 1.83, 95% CI 1.03-3.24). However, after excluding studies deviating from HWE, we found the association disappeared among European descendants (G versus A: OR - 1.61, 95% CI- 0.94-2.75; GG versus AA: OR - 2.39, 95% CI - 0.73-7.79; GG versus GA+AA: OR - 1.80, 95% CI - 0.80-4.08). Significant heterogeneity was found in the overall analyses even after removing studies deviating from HWE. However, some of the heterogeneity can be eliminated through ethnicity-specific analyses, which is observed in the studies conducted in Caucasian descendants. Among other subgroup analyses, only studies conducted in African descendants achieved improved heterogeneity after excluding studies deviating from HWE (Table 2). Results from Begg’s and Egger’s test suggested that publication bias may present in the studies investigating the association between the IL-10-1082G/A polymorphism and TB risk among Asian population (Table 2).
Meta-analysis of interleukin (IL)-10-1082G/A polymorphism and tuberculosis risk according to a study from China.
Quantitative synthesis of IL-10-819C/T
As shown in Table 3, the pooling analysis of total included studies indicated no association between IL-10-819C/T polymorphism and TB risk (C versus T: OR - 0.96, 95% CI - 0.91-1.01; CC versus TT: OR- 0.95, 95% CI - 0.85-1.07; CT versus TT: OR- 1.03, 95% CI - 0.86-1.23; CC+CT versus TT: OR - 0.98, 95% CI - 0.84-1.14; CC versus CT+TT: OR - 0.90, 95% CI - 0.78-1.05). Excluding studies deviating from HWE did not affect the results (C versus T: OR - 0.97, 95% CI - 0.92-1.03; CC versus TT: OR - 0.98, 95% CI - 0.86-1.11; CT versus TT: OR - 0.98, 95% CI - 0.89-1.08; CC+CT versus TT: OR - 0.96, 95% CI - 0.88-1.06; CC versus CT+TT: OR- 0.96, 95% CI - 0.87-1.05). In the subgroup analyses by ethnicity, IL-10-819C/T polymorphism was associated with decreased TB risk among Asian descendants under 3 genetic models (C versus T: OR - 0.89, 95% CI - 0.82-0.96; CC versus TT: OR - 0.81, 95% CI - 0.67-0.97). The association was still significant after excluding studies deviating from HWE (C versus T: OR - 0.88, 95% CI - 0.81-0.97; CC versus TT: OR - 0.79, 95% CI - 0.64-0.97; CC+CT versus TT: OR - 0.87, 95% CI - 0.77-0.98; CC versus CT+TT: OR - 0.82, 95% CI - 0.68-0.98). While no significant association was found in other ethnicity in all tested models (Table 3). The heterogeneity was significant among some genetic models in the overall and subgroup analyses. However, most of heterogeneity was eliminated after excluding studies deviating from HWE (Table 3). Begg’s and Egger’s test suggested that publication bias was not apparent in all analyses concerning IL-10-819C/T polymorphism (Table 3).
Meta-analysis of interleukin (IL)-10-819C/T polymorphism and tuberculosis risk as found in a study from China.
Quantitative synthesis of IL-10-592C/A
The main results of the relationship between IL-10-592C/A polymorphism and TB risk are presented in Table 4. Similar to the other 2 genetic site polymorphism, the overall analysis of IL-10-592C/A polymorphism, including all ethnic groups, showed no significant association with TB risk (C versus A: OR - 0.97, 95% CI - 0.85-1.11; CC versus AA: OR - 0.89, 95% CI - 0.68-1.17; CA versus AA: OR - 0.94, 95% CI - 0.77-1.14; CC+CA versus AA: OR - 0.94, 95% CI - 0.76-1.16; CC versus CA+AA: OR - 0.94, 95% CI - 0.78-1.13). Removing the studies deviating from HWE yielded almost similar results (C versus A: OR - 1.00, 95% CI - 0.87-1.16; CC versus AA: OR - 0.94, 95% CI - 0.71-1.26; CA versus AA: OR - 0.93, 95% CI 0.76-1.13; CC+CA versus AA: OR - 0.94, 95% CI - 0.76-1.17; CC versus CA+AA: OR - 0.98, 95% CI - 0.80-1.21). Subgroup analyses were also performed by ethnicity. Decreased risk was observed among Asians in 3 genetic models (C versus A: OR - 0.79, 95% CI - 0.65-0.97; CC versus AA: OR - 0.57, 95% CI - 0.42-0.76; CC versus CA+AA: OR - 0.66, 95% CI - 0.51-0.86). When removing the studies deviating from HWE in Asians, we found significant association in all 5 genetic models (C versus A: OR - 0.74, 95% CI - 0.65-0.85; CC versus AA: OR - 0.55, 95% CI - 0.41-0.75; CA versus AA: OR - 0.73, 95% CI - 0.60-0.89; CC+CA versus AA: OR - 0.69, 95% CI - 0.58-0.83; CC versus CA+AA: OR - 0.66, 95% CI - 0.51-0.86). While increased risk among Europeans in 2 genetic models (C versus A: OR - 1.31, 95% CI - 1.02-1.67; CC versus AA: OR - 1.88, 95% CI - 1.05-3.37) and Caucasian in 2 genetic models (C versus A: OR - 1.23, 95% CI - 1.06-1.42; CC versus CA+AA: OR - 1.45, 95% CI - 1.14-1.83) were observed in ethnicity-specific analyses. The results cannot be changed after excluding studies deviating from HWE among Europeans (C versus A: OR - 1.31, 95% CI - 1.02-1.67; CC versus AA: OR - 1.88, 95% CI - 1.05-3.37) and Caucasian (C versus A: OR - 1.25, 95% CI - 1.08-1.45; CC versus CA+AA: OR - 1.48, 95% CI - 1.16-1.89). No significant association was found among other populations (Table 4). Obvious heterogeneity was found in overall analyses. However, removing studies deviating from HWE, all subgroups exhibited homogeneity in all genetic models (Table 4). Begg’s and Egger’s test showed no evidence of publication bias in all analyses concerning IL-10-592C/A polymorphism (Table 4).
Meta-analysis of interleukin (IL)-10-592C/A polymorphism and tuberculosis risk as found in a study from China.
Discussion
At present, 3 meta-analyses have been conducted to assess the association between IL-10 polymorphisms and TB risk. Due to a relatively smaller study sample size, it is difficult for these previous studies to arrive at solid conclusions. Pacheco et al,58 who conducted the first meta-analysis including only 8 studies did not find any significant association between IL-10-1082 G/A polymorphism and TB risk, whereas Zhang et al59 reported an increased risk of TB in Europeans with IL-10-1082 in the genetic model (GG versus AA+AG). Recently, Liang et al60 conducted a meta-analysis, and provided evidence that IL-10-1082G/A polymorphism was associated with TB risk in Europeans and Americans, and IL-10-819T/C and -592A/C polymorphisms could be risk factors for TB in Asians. Although the latter 2 meta-analyses,59,60 which included 18 European, and 31 American studies have to some extent, amplified the power of test, they did not perform sensitivity analysis by excluding studies deviating from HWE to confirm the stability of results. Therefore, we carried out this meta-analysis on the basis of 37 eligible case-control studies to comprehensively, and rigorously evaluate the association between IL-10 polymorphisms and TB risk.
In terms of the IL-10-1082 G/A polymorphism, the results in pooling all studies showed no significant associations between the IL-10-1082 G/A polymorphism and TB risk. In the subsequent subgroup analysis, our initial pooling analyses found that this polymorphism in European group indicated an increased TB risk in 3 genetic models (allelic model, homozygote comparison, and recessive model), which is in consistency with Zhang et al’s59 and Liang et al’s60 meta-analysis. However, after excluding studies deviating from HWE during sensitivity analysis, our research failed to replicate this positive finding. As including studies deviating from HWE may lead to false-positive finding, those positive results should be interpreted with caution. Meanwhile, it is worth emphasizing that our meta-analysis showed an association between IL-10-1082 G/A polymorphism and TB risk in Caucasian descendants without between-study heterogeneity, which is also in consistency with the research conducted by Liang et al.60 In view of the relatively small sample size of people were included in the pooling analysis of Caucasian subgroup, additional large studies are warranted to validate this finding.
Although pooled results of all the included studies demonstrated that IL-10-819C/T polymorphism has no substantial effect on the occurrence of TB, the finding that this polymorphism in 4 genetic models (allelic model, homozygote comparison, dominant model, and recessive model) was linked to protective effect on TB in Asian population was worth to be noted. It is a relatively solid result without between-study heterogeneity, and is also in accordance with the previous meta-analysis conducted by Liang et al.60 While no significant association was found in other ethnicity in all tested models, which was also consistent with previous reports.59,60 The IL-10-592C/A polymorphism was not linked to any obvious risk or protective effect on TB in all population. Interestingly, subgroup analyses showed that IL-10-592C/A polymorphism was associated with decreased susceptibility to TB in the Asian group, and increased susceptibility in European and Caucasian group, which were not reported by former meta-analyses.58-60
The reason why different association was observed in different ethnicity remained unclear. One possible explanation is the differences in the frequencies of the polymorphisms among diverse ethnicity, which may partly give rise to heterogeneity. Meanwhile, gene-gene interactions along with environmental factors could also contribute to the complexity of genetic effect. Thus, most often, variation in single genetic locus is insufficient to predict risk of disease.
The strength of our meta-analysis included a large sample size, comprehensive search strategy, and no language restrictions. As a result, our meta-analysis has more power to detect differences, and allow subgroup analysis. Additionally, we performed sensitive analysis by excluding studies deviating from HWE to confirm the stability of the results, which then may lead to more reliable conclusions.
Some limitations in this meta-analysis should also be addressed. Firstly, publication bias was apparent in the IL-10-1082 G/A genetic site under some genetic models among Asians. Secondly, some detailed information, such as HIV status, or TB severity was unavailable in most studies, which limited our further evaluation by performing stratified analysis according to those confounding factors. Thus, future studies should provide more information, such as the HIV status and TB severity, to promote further assessing of the association between IL-10 genetic polymorphism and TB risk. Thirdly, the small sample sizes in some subgroup analyses may not be the representative of the population. Therefore, the relationship between IL-10 genetic polymorphisms and TB should be confirmed in future studies.
In conclusion, this meta-analysis involving 18,553 participants evaluated the relationship between IL-10-1082/-819/-592 polymorphisms and risk of TB, which provided evidence that IL-10-819C/T and -592C/A polymorphisms are associated with TB susceptibility in Asians, and IL-10-592C/A polymorphism is related to TB susceptibility among Europeans, and IL-10-1082 G/A, -592C/A polymorphisms may be in association with TB susceptibility in Caucasian. Large-scale studies are warranted to be undertaken to confirm those relationship. Future studies should provide more information, such as the HIV status and TB severity, to promote further assessing of the association between IL-10 genetic polymorphism and TB risk.
Footnotes
Disclosure. Authors have no conflict of interests, and the work was not supported or funded by any drug company. This research was supported by the Science and Technology Department of Sichuan Province (grant no. 2012SZ0126), and National Natural Science Foundation of China (grant no. 81072432 and 81301400).
- Received November 2, 2014.
- Accepted February 1, 2015.
- Copyright: © Saudi Medical Journal
This is an open-access article distributed under the terms of the Creative Commons Attribution-Noncommercial-Share Alike 3.0 Unported, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.