Abstract
Objectives: To explore the risk factors, the prevalence rate, and gene types of extended-spectrum beta-lactamase (ESBL)-producing bacteria as the causative agents of infection at King Abdulaziz Specialist Hospital (KAASH), Taif, Kingdom of Saudi Arabia.
Methods: This was a retrospective study conducted during the period between February 2017 and January 2018. All samples obtained from the KAASH were analyzed. The MicroScan Walkaway System, bacteriological examination and double disk synergy tests were used to detect ESBL-producing bacteria. To identify ESBL genes, the polymerase chain reaction (PCR) technique was used.
Results: The ESBL phenotype was detected in 351 of 1151 isolates (30.5%); Escherichia coli (E. coli) (62.7%) and Klebsiella pneumoniae (K. pneumoniae) (23.6%) were the most prevalent. The highest proportion of ESBL specimens was found in urine (62%.5), and these organisms were mainly isolated from the female medical ward (20.2%). Based on the statistical analysis, lung diseases, renal diseases, diabetes and heart diseases contributed to the spread of ESBL infections. Amikacin, imipenem, meropenem and tigecycline were found to be effective in overcoming ESBL infections; however, these antibiotics may be inappropriate for new strains of K.pneumoniae. The distribution of the blaCTX-M gene was high (87%), compared with blaTEM (74.9%) and blaSHV (29.4%).
Conclusion: These data provide new epidemiological information about the prevalence of ESBL-producing organisms among patients in KAASH, Taif, Saudi Arabia. In addition, this study identified the clonal nature of isolated E.coli and K.pneumoniae.
Today, one challenge faced by the World Health Organization (WHO) is extended-spectrum beta-lactamase (ESBL)-producing bacteria. Rates of ESBL production among Enterobacteriaceae range between 6-38% in Kingdom of Saudi Arabia, with the highest frequency recorded in the central regions.1 Excessive use of third-generation cephalosporins, which are commonly used as an empiric and definitive therapy in hospitals, are expected to trigger the emergence and spread of ESBL producers.2
There are several risk factors that may contribute to the prevalence of ESBL infection among patients and healthcare workers. Studies aimed at identifying these factors, and the extent of their association with ESBL-producing bacteria, have largely agreed on the contributions of certain factors, such as hospital admission and the previous use of antibiotics, but have conflicting results with respect to the impact of other factors such as gender, age, and chronic disease (namely, diabetes and renal disease). Therefore, factors linked with the spread of ESBL infection remain controversial issues and require further research to determine the relationship between these factors and the acquisition of ESBL infection.
Extended-spectrum beta-lactamases-producing bacteria have the ability to secrete common enzymes, such as blaCTX-M, blaTEM and blaSHV, and almost 450 variants of these enzymes have been detected.3 This is undoubtedly due to significant advancements in gene detection methods, such as polymerase chain reaction (PCR), that provides very accurate results. Extended-spectrum beta-lactamases producing organisms break the beta-lactam ring, which leads to resistance to antimicrobials, such as cephalosporins and monobactams.4 In addition, extensive use of these drugs leads to mutations of ESBL genes and the emergence of ESBL infection. The remarkable proliferation of such bacteria globally is causing an increase in the proportion of infected patients in the community and hospitals, as well as higher mortality rates. Various studies have indicated that the spread of such bacteria, which are capable of secreting these enzymes, might be related to the geographic region. For instance, in Saudi Arabia, the prevalence of blaSHV in Klebsiella pneumoniae (K. pneumoniae) isolates was approximately 78% in Al-Ahasa and less than 14% in Mecca hospitals.5,6
According to several researches, blaCTX-M-15 is predominant, compared with other genes.7 Clonal proliferation and genetic elements have been considered causes of this spread.8 This retrospective study aims to identify the risk factors of infection caused by ESBL-producing bacteria at the KAASH, Taif, Kingdom of Saudi Arabia and to identify the different types of ESBL enzymes that are found using PCR methods.
Methods
This was a retrospective study and the isolates analyzed were from the KAASH in Taif, Kingdom of Saudi Arabia which has a 500-bed capacity. It is a referral hospital and serves as a teaching hospital for Taif University Medicine College, Taif, Saudi Arabia. This study was conducted between February, 2017 and January, 2018. Urine, blood, sputum, swabs and other clinical specimens were cultured on blood agar and MacConkey agar (Saudi prepared media laboratory [SPML]). Patients with gram-negative bacteria (GNB) infections were recorded. Only one sample from each patient was considered. Identification of the organisms was performed using the Microscan Walkaway system (Beckman Coulter, USA), according to the manufacturer’s instructions. Other tests such as analytic profile index (20EAPI-20E) and E-test (bioMérieux, France), and double disc synergy test (DDST) were also performed if needed.
The ESBL production was performed according to the Clinical and Laboratory Standards Institute (CLSI) guidelines of 2012.9 Mueller-Hinton agar (SPML) plates were inoculated with the text organisms. Ceftazidime and co-amoxiclav discs were placed 30 mm apart (centre to centre) and incubated at 37°C for 24 hours. Enhancement of the inhibition zone in the area between the discs indicated a positive result. The E-test (bioMerieux, France) was used to confirm the ESBL result. The specimen was spread onto a Mueller-Hinton agar plate. If the ratio of the minimum inhibitory concentration of co-amoxiclav to that of ceftazidime was more than 8, the result was considered positive. Routine susceptibility was determined using the disc-diffusion method on Mueller-Hinton agar, as recommended by the CLSI. To ensure quality control, Escherichia coli (E. coli) (ATCC 25922) and K. pneumoniae (ATCC 700603) ESBL-producing strains were used as controls for the antimicrobial susceptibility tests.
A genomic DNA purification kit (Jena Bioscience GmbH, Germany) was used to purify ESBL-producing E. coli (n=209, 59.5%) and K.pneumoniae (n=90, 25.6%) isolates. The PCR amplification of blaCTX-MF was performed using the following primers: (5′-TTAGGAARTGTGCCGCTGYA-3′) and blaCTX-MR was: 5′-CGATATCGTTGGTGGTRCCAT-3′ with an amplicon size of 688bp.10 Primers used for blaTEM-C was: 5′-ATCAGCAATAAACCAGC-3′ and blaTEM-H was: 5′-CCCCGAAGAACGTTTTC-3′ with an amplicon size of 516bp.11 Primers used for blaSHV-F was: 5′-AGGATTGACTGCCTTTTTG-3′ and blaSHV-R was 5′-ATTTGCTGATTTCGCTCG-3′ with an amplicon size of 392pb.11 For amplification, 2 µl template DNA (around 50 ng) was added to 23 µl reaction mixture, which contained 12.5 µl GoTaq green master mix (Promega, USA), 0.5 µl of each primer and 10 µl nuclease-free water. A C1000 Touch thermal cycler (Bio-Rad, Germany) was used to complete the PCR reaction as follows: denaturation at 94°C for 5 minutes was followed by 35 cycles of denaturation at 94°C for 45 seconds, then annealing for 45 seconds (at 50°C for blaTEM and blaSHV and at 60°C for blaCTX-M), extension at 72°C for 60 seconds and a final extension at 72°C for 7 minutes. The polymerase chain reaction amplicons were separated by electrophoresis on a 1.5% agarose gel in 1X TAE (Tris/Acetate/EDTA) buffer containing 0.5ug/ml ethidium bromide. The last step was photographed under an ultraviolet lamp (Bio-Rad Gel Doc 2000 device).
Statistical analysis
Microsoft Software 2010 was used in the analysis of the results, which are presented as frequencies or percentages. This software was also used to determine the p-values of different characteristics of patients who were infected with ESBL.
Results
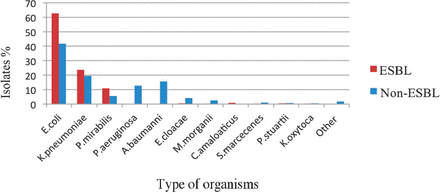
A total of 351 ESBL-producing bacteria were isolated from 1151 GNB (inpatients 87.7% and outpatients 12.3%). The majority of the ESBL-producing isolates were E. coli (220/351, 62.7%), followed by K. pneumoniae (83/351, 23.6%) and Proteus mirabilis (38/351, 10.8%). Other microbes that can secrete these enzymes such as Enterobacter cloacae, Morganella morganii, Serratia marcescens, Klebsiella oxytoca, Providencia stuartii and Citrobacter amalonaticus were less widespread in the hospital environment (10/351, 2.8%) (Figure 1).
Distribution of extended-spectrum beta-lactamase (ESBL) and non-ESBL isolates according to species. E. coli - Escherichia coli, K. pneumoniae - Klebsiella pneumoniae, P. mirabilis - Proteus mirabilis, P. aeruginosa - Pseudomonas aeruginosa, A. baumannii - Acinetobacter baumannii, E. cloacae - Enterobacter cloacae, M. morganii - Morganella morganii, C. amalonaticus - Citrobacter amalonaticus, S. marcescens - Serratia marcescens, P. stuartii - Providencia stuartii, K. oxytoca - Klebsiella oxytoca.
Extended-spectrum beta-lactamase isolates were mainly recovered from urine in high percentages of 62.1% (218/351). Other ESBL isolates were recovered from swabs (49, 14%), followed in decreasing order by blood (36, 10.2%), sputum (32, 9.1%) and other (16, 4.6%).
Table 1 compares the percentages of ESBL among males, females, and different age groups. Remarkable differences between males and females were found, with ESBL infection presenting in more females than males (59.8%, 210/351). Furthermore, females presented almost the same percentage of non-ESBL producers as ESBL producers at 54.8%. The proportion of females aged ≥80 years among the ESBL and non-ESBL producers was lower than that of males in the same age bracket and was 6% and 8%. For males, the number of non-ESBL producer bacteria outnumbered females by roughly 2-one in the age groups of 0-9 and 10-19 years old (13%, 47/362 and 6.6%, 29/438). In addition, the spread of ESBL infection among males aged 10-19 showed the lowest level among either gender with nearly 2% (6/351).
Age distribution of patients with ESBL producers and non-ESBL.
Extended-spectrum beta-lactamase infection was the most common in the female medical ward, with approximately 21% of the total isolates. The spread of ESBL-producing bacteria among patients in the male surgical ward was also high (17.7%), compared with other wards. In contrast, ESBL infection was less spread among patients in the male medical ward-high dependency unit and occupational therapy and had, the same rate of 0.6%. The male medical ward, female surgical ward and intensive care unit (ICU) were other sections in which ESBL infection spread among patients, with about 13% (Table 2).
Distribution of ESBL-producing and non-ESBL bacteria according to hospital wards.
The total number of non-Saudi patients was 24, and all of these patients were from Asia or Africa (13.5%, 155/1151). The ESBL infection accounted for 9.7% of the total isolates (34/351) and most of these patients were Yemeni (29.4%) or Pakistani (14.7%).
To determine the risk factors that might lead to ESBL infection among patients, the most important characteristics of patients were divided into several variables (Table 3). These variables included age, gender, underlying diseases and the use of certain antibiotics (such as gentamicin, nitrofurantoin and norfloxacin). In addition, the number of patients with ESBL infection was associated with the presence or absence of these characteristics. For example, kidney diseases accounted for roughly 28% (98/351) of the total number of patients infected with ESBL, whereas patients with non-ESBL were 16.9% (135/800).
Potential risk factors and clinical traits for ESBL-producing bacterial infections.
Several clinical features are associated with the respiratory system, such as community-acquired pneumonia and chronic obstructive pulmonary disease (COPD). These diseases accounted for around 5% of patients with an ESBL infection, and were reported among approximately 3% of the patients with non-ESBL infections. This finding indicates that there is an important relationship between some respiratory diseases and ESBL infection (p=0.04). Furthermore, heart diseases included 6 variables in this study. Complete heart disease, post-cardiac arrest, stroke, hypertension, ischemic heart disease and atrial fibrillation indicated an important affiliation with ESBL infection (p=0.003).
For gender, the proportion of women and men with ESBL infections indicate that gender is a non-significant factor for acquiring this infection (p=0.7 and p=0.2). In addition, age did not have an effect on ESBL infection among different age groups (p>0.05). It was clear from the results that patients with diabetes were most likely to acquire ESBL than were other patients (p=0.02). However, there was no significant link between ESBL infection and other comorbid conditions, such as central nervous system and cancer (p=1 and p=0.8). The antibiotics, gentamicin, nitrofurantoin and norfloxacin did not have a significant connection with ESBL infection (p>0.05). In this study, some antibiotics were used to test the sensitivity of ESBL-producing E.coli and K.pneumoniae, which accounted for around 86% of the total ESBL-producing bacteria. The penicillin and cephalosporin groups failed to treat such organisms, and had a resistance percentage of 100% (Table 4). However, several antimicrobials had a large effect in overcoming ESBL infections with rates ranging from 80-100%, such as amikacin, imipenem, meropenem and tigecycline. In addition, some antimicrobials had the ability to treat ESBL-producing E.coli at a greater percentage than ESBL-producing K.pneumoniae. For instance, nitrofurantoin was a significant drug for curing E.coli (susceptibility [S]=95%); however, it was not an appropriate choice for patients infected with K.pneumoniae (S=40%).
Antimicrobial susceptibility patterns of ESBL-producing E.coli and K.pneumoniae isolates.
Both organisms were able to resist norfloxacin and ciprofloxacin with a high rate for E.coli (77% and 71%) and a low percentage for K.pneumoniae (32% and 48%). Furthermore, a combination of medication, such as piperacillin/tazobactam, was effective for treating ESBL-producing E.coli at 87%, yet was less effective for treating ESBL infection caused by K. pneumoniae at 75%.
Amoxicillin/clavulanate is another example drug combination that was used to treat ESBL infection; however, the results of susceptibility of ESBL-producing E.coli and K. pneumoniae indicated that this antimicrobial was not suitable for such infection.
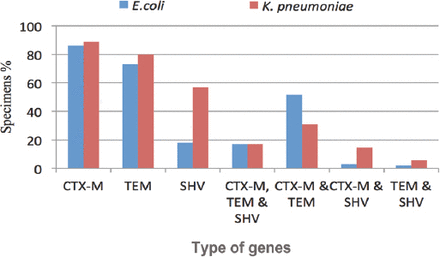
The results indicated that the level of blaCTX-M secretion was high compared with blaTEM and blaSHV (87%, 75%, and 29%). Klebsiella pneumoniae could produce 2 types of enzymes (blaCTX-M and blaTEM) at a higher rate than E.coli (Figure 2). The blaSHV gene production by K.pneumoniae (57%) was also about 3 times greater than the number of E.coli. Also, blaCTX-M, blaTEM, and blaSHV were secreted together, equally by both organisms at nearly 17%. Escherichia coli released both blaCTX-M and blaTEM more so than K.pneumoniae, at 51%.
The extended-spectrum beta-lactamase (ESBL) genes distribution among Escherichia coli, and Klebsiella pneumoniae.
For secreting blaCTX-M and blaSHV together, K.pneumoniae was able to secrete these enzymes with a higher percentage than E.coli at 13% and 3%. The blaTEM and blaSHV genes were the lowest of that ESBL enzymes that were produce by K.pneumoniae and E.coli at the same time (<6%). In general, K.pneumoniae had the ability to secrete the 3 enzymes in higher amounts than E.coli, whether they were together or alone, except for the production blaCTX-M and blaTEM simultaneously.
Discussion
Over the past 10 years, ESBL infections have spread throughout the different regions of Kingdom of Saudi Arabia with a high rate of 36%.12 Within the Western Kingdom of Saudi Arabia region, high ESBL prevalence has been reported among patients in Mecca (30%) and in Jeddah (20%).6,13 In the present study, the prevalence of ESBL producers among gram-negative bacteria was 351 of 1151 (30.4%), which corresponds to the findings of Al Mously et al,14 in 2016 (27.4%). However, different results were reported for ESBL infection in other areas of Kingdom of Saudi Arabia, such as in the Central and the Eastern regions, with rates of 6.3% and 23%.15,16
In this study, infection with ESBL-producing E.coli (62.7%) was the most common, followed by K.pneumoniae (23.6%), P.mirabilis (10.8%) and others with lower rates (2.8%). Similar findings were reported by Elhassan et al,11 which revealed that E.coli and K.pneumoniae were detected at high rates of 77.6% and 22.4%. Although P.aeruginosa was recorded as an ESBL-producing bacterial species with a prevalence of 25.9% in the Western region (Mecca) by Ahmed et al,17 this might have been related to Umrah or Hajj; all P.aeruginosa specimens in our study were non-ESBL producers.
In this study, 59.8% of positive ESBL samples were from females, compared with 40.2% from males, which is comparable to the results published by Tham et al,18 2013 (women, 77%), but differs from other studies that found that the majority of ESBL patients were male.19 The proportion of patients aged <60 years was greater than that of patients aged ≥60 years at 57.3% and 42.7%. However, there was no significant association between age and ESBL infection (p>0.05). The same finding was reported by Saudi researchers and others.12,20 In addition, non-Saudis who were infected with ESBL producers accounted for 9.7% of positive samples, and most of these patients were Yemeni (29.4%), followed by Pakistani (14.7%). These results might be due to the large proportion of these nationalities in Taif, Kingdom of Saudi Arabia. In contrast, the study by Zaman et al,13 2015 (in Mecca) indicates that the distribution of Saudi patients are less than that of non-Saudi patients: 32.3% and 67.7%. These results might have been affected by the population diversity that characterizes this region.
Regarding specimen type, urine accounted for the largest proportion of specimens positive for ESBL producers (218/351, 62.1%). Furthermore, ESBL producers were isolated from swabs (14%), blood (10.3%) and sputum (9.1%). Other researchers showed similar findings, with urine accounting for approximately 60% of the samples.11,12 Extended-spectrum beta-lactamase producers were primerly isolated from the female medical ward (20.2%), male surgical ward (17.7%), and male medical ward and female surgical ward (13.4%) and ICU (12.3%). These findings differ from local and global studies in which the highest percentages of ESBL producers were isolated from the ICU.7,21
In the present study, the lowest rates of ESBL infections were in female patients aged 90-99 years (3.3%), while the highest percentages were from female patients aged 30-39 years (18%). A study in 2014 reported that patients aged ≤18 years were less likely to be infected with ESBL bacteria.15 During the period 2006-2011, Kassakian and Mermel studied 321 ESBL specimens and found that the age of patients was >65 years and that approximately 74% were female.22 However, older age and gender were not significantly correlated with ESBL infection in the present study (p>0.05), and these results correspond with data reported by other investigations (p=0.3 and p=0.4).12,23
Kidney dysfunction is a serious disease that, is associated with ESBL-producing Enterobacteriaceae. Several studies indicate that the prevalence of ESBL-producing E.coli and K.pneumoniae among patients with renal disease was noteworthy.24 In this research, the spread of ESBL-producing strains in patients caused significant renal dysfunction during various stages of the disease and, resulted in kidney failure (p=0.0001). These findings were consistent with those previously reported for the King Khalid University Hospital in Riyadh, Kingdom of Saudi Arabia.25
Several studies have reported diabetes as a risk factor for ESBL infection. In 2016, for example, Al Yousef et al,26 isolated 296 ESBL samples and found that diabetes was a risk factor associated with ESBL infection. In addition, Rahim et al,27 found that diabetes was significantly linked to ESBL infection. The current study reveals that diabetes is significantly associated with ESBL infection (p=0.02). However, a retrospective study performed in Turkey in 2013, showed that there was no link between diabetes and ESBL infection (p=0.3).28 Based on multivariate analysis, there were several characteristics related to lung diseases, such as COPD and respiratory disease syndrome (RDS). The percentage of lung disease cases with ESBL-producing bacteria was greater than non-ESBL at 5.4% and 2.8%. This finding reveals the important relationship between such diseases and ESBL infection (p=0.04). Other studies by Blom et al,29 and Vijayakanthi et al,30 confirmed that COPD and RDS were underlying illnesses associated with ESBL infection.
Heart failure/heart disease causes and ESBL infection have been reported in abundant scientific research. Some of these studies did not find an important connection between such diseases and ESBL organisms.18 However, the study by Quan et al,31 concluded that heart failure may be associated with the prevalence of ESBL K.pneumoniae in hospitals (p=0.001). In this investigation, several comorbidities related to heart diseases showed a significant link to ESBL-positive Enterobacteria (p=0.003). In general, the current study indicates that piperacillin/tazobactam, amikacin and nitrofurantion, have the ability to treat both ESBL and non-ESBL infections. These results correspond to those published by Khalid et al,32 who recommended the use of nitrofurantion as an oral drug against ESBL producers and other urinary pathogens. In addition, several drugs retain the ability to overcome ESBL-producing bacteria. For instance, in this data, imipenem, meropenem and tigecycline showed the capacity to cure a large range of ESBL bacteria.
Extended-spectrum beta-lactamase-producing bacteria carry highly diverse enzymes that include blaCTX-M, blaTEM, blaSHV, blaOXA, blaPER, blaVEB and blaBES.33 The first and the second genes were more abundant than the other genes. Certain factors contribute to the prevalence of ESBL infection, such as society, health organizations and the economics of different countries.34 Different studies have recorded high rates of ESBL infection in India (80%), with a large population and inadequate quality of life, and Egypt (70%), compared with 2.5% in Sweden and and 5% in America. In the Middle East, this percentage varies from 16% in Lebanon to 26% in Kuwait and 38% in Kingdom of Saudi Arabia.34
Furthermore, the spread of a specific type of bacteria plays an important role in increasing resistance to specific types of antibiotics. For example, blaCTX-M is associated with E.coli in many cases, while blaTEM and blaSHV are linked to K.pneumoniae, and both display resistance to the third-generation cephalosporins.35 The blaOXA gene appears in P.aeruginosa, which has the ability to resist cloxacillin and oxacillin, but is more sensitive to the cephalosporins group.36 In this study, blaCTX-M was predominant with a high rate of 87%, followed by blaTEM (74.9%) and blaSHV (29.4%). Sharma et al,37 shared this finding with, 83% for blaCTX-M and 67% for blaTEM.
Some studies have shown that certain bacteria are able to secrete 2 or 3 enzymes. For example, a study consisting of 81 ESBL-producing E.coli and 21 K.pneumoniae found that certain strains of E.coli have the ability to produce blaCTX-M, blaTEM and blaSHV, accounting for 13% of total secreted enzymes, while other strains of the same organism released blaTEM and blaSHV at 2.5%. Klebsiella pneumoniae had the same ability to produce 2 enzymes (blaTEM and blaSHV) and 3 enzymes (blaCTX-M, blaTEM and blaSHV), at 19% of total isolates.6 Other local investigation revealed that both blaSHV and blaTEM were released at the low level of 3.5%.11 In the present study, both blaTEM and blaSHV were detected in 3% of isolates, while 17% of the isolates carried all 3 genes together, blaCTX-M, blaTEM and blaSHV. The distribution of blaSHV depends on the rate of ESBL infection caused by K.pneumoniae. A study by Dehshiri et al,38 on the occurrence of ESBL-producing K.pneumoniae in patients with urinary tract infection concluded that the prevalence of blaSHV was very high (85%), compared with that of blaCTX-M and blaTEM at 30% and 16%. In the present investigation, the prevalence of blaSHV was greater in K.pneumoniae (56.7%) than E.coli (17.7%). The high prevalence of these enzymes may contribute increasing antimicrobial resistance among Enterobacteriaceae. These resistant strains are selected by overuse and misuse of antibiotics in our locality because of the high prevalence of antibiotic misuse which has been reported in Taif, Kingdom of Saudi Arabia as well as other regions in Kingdom of Saudi Arabia.39
Study limitation
The PCR technicque was performed to detect genotypes of 2 organisms, E.coli and K. pneumoniae. In addition, the detected blaTEM and blaHSV genes were not sequences.
In conclusion, this study highlights an important link between ESBL prevalence and certain factors such as renal disease, respiratory diseases, heart diseases and diabetes. In addition, the uncontrolled use of antibiotics has increased the resistance of ESBL-producing bacteria. Fortunately, various drugs, that include amikacin, imipenem and meropenem, retain the ability to overcome ESBL infection. However, the presence of K.pneumoniae in hospitals or in the community contributes to increase the distribution of the blaSHV gene, which will create more suffering for patients. Additional studies are needed to determine other factors that may be associated with ESBL infection and to determine the appropriate solutions to reduce the effects of these factors on the spread of this infection.
Acknowledgment
The authors would like to thank DSR for technical and financial support. We also would like to thank the staff of the Research Unit and the Laboratory Department at King Abdulaziz Specialist Hospital in Taif, Kingdom of Saudi Arabia for their kind assistance in the data collection.
Footnotes
Disclosure. This study was funded by Deanship of Scientific Research (DSR) under grant no. G-517-130-38, at King Abdulaziz University, Jeddah, Kingdom of Saudi Arabia under grant no.
- Received August 8, 2018.
- Accepted October 31, 2018.
- Copyright: © Saudi Medical Journal
This is an open-access article distributed under the terms of the Creative Commons Attribution-Noncommercial-Share Alike 3.0 Unported, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.