Abstract
Objectives: To identify the impact of Lipocalin-2 (LCN2) gene polymorphisms on breast cancer patients in Jeddah, Saudi Arabia.
Methods: It is a case control study in which blood samples of participants from Medical Reference Clinics and King Abdulaziz University Hospital in Jeddah, Saudi Arabia have been taken between 2014 and 2016. This study recruited 128 participants (50% control, 50% patients) and used Tetra-Primer amplification-refractory mutation system-polymerase chain reaction method for the detection of missense SNP (rs11556770). The study measured LCN2 plasma protein expression by enzyme-linked immunosorbent assay technique.
Results: The results have shown that 100% of the genotypes were normal allele (G/G). In contrast, the plasma level of LCN2 was considerably elevated among patients as compared to control (p=0.001), and higher in invasive ductal carcinoma patients (p=0.001). The LCN2 protein expression in plasma level was significantly elevated among patients, particularly who demonstrated invasive ductal carcinoma.
Conclusion: There is no significant relationship between breast cancer patients and LCN2 gene polymorphisms (rs11556770).
Breast cancer is one of the severe cases of malignancy and is a major cause of mortality across the globe. This pathologic condition accounted for one million cases diagnosed, and approximately half million deaths yearly.1,2 Breast cancer has now become an alarming public healthcare issue within Saudi Arabia. Breast cancer has made approximately 19.9% of all cancer cases in Saudi Arabia during 2012. This data was recorded among women over the age of 52 years.3 It is statistically projected that one in every 8 women in the United Staes of America is affected by invasive cancer.4 Existing literature suggested that the common genes, which cause breast cancer include; BReast CAncer susceptibility gene (BRCA)1 and BRCA2;5,6 even though, there are increased Lipocalin-2 (LCN2) levels in the breast cancer cells among women. Incidence rates vary according to region, being lower in developing nations and higher in developed countries.2 It has become evident that LCN2 is over-expressed in cancers, concerning varied histological origin. It may also facilitate tumorigenesis by promoting growth, metastasis, and survival.7 Lipocalin-2 (LCN2) located on the 9th chromosome cytogenetic band 9q43.11 is associated with lipocalin (NGAL) and is known as the neutrophil gelatinase8 on the 9th chromosome cytogenetic band 9q43.11. It is composed of 178 different amino acids, which have 863 bp coding region, encompassing seven axons and 6 introns.9 Its abnormal expression is observed in malignant human cancers that plays a significant part in tumor metastasis. The overexpression of LCN2 is responsible for enhancing the invasion abilities and migration of 4T1 cells resulting in metastasis. The promotion of metastasis occurs by inhibiting the P13K/Akt pathway. Unlike, cell lines derived from benign breast cancers, increased amounts of Neutrophil Gelatinase Associated Lipocalin (NGAL) lipocalin are secreted from cell lines that are derived from highly metastatic breast cancer.10 The expression of lipocalin is restricted to the breasts only. The roles of lipocalin have not been shown clearly in the formation of breast cancer; however, it is observed to correlate with breast cancer.11,12 According to Candido et al,13 LCN2 has a significant role in tumor development at early stages. Epithelial cells were found, containing a higher amount of LCN2 gene expression; hence, LCN2 participates aggressively in promoting cancer growth in breast cancer cells.14 Lipocalin-2 located on the 9th chromosome cytogenetic band 9q43.11 is associated with lipocalin (NGAL) and is known as the neutrophil gelatinase8 on the Chromosome 9 cytogenetic band 9q43.11. It is composed of 178 different amino acids, which have 863 bp coding region, encompassing 7 axons and 6 introns.9 Its abnormal expression is observed in malignant human cancers that plays a significant part in tumor metastasis. The overexpression of LCN2 is responsible for enhancing the invasion abilities and migration of 4T1 cells resulting in metastasis. The promotion of metastasis occurs by inhibiting the P13K/Akt pathway. Other researchers have found higher levels of LCN2 among patients with endometrial cancer.15-18
Therefore, the aims of this study is to investigate the impact of LCN2 gene polymorphisms (SNP rs11556770) on breast cancer patients using Tetra-Primer amplification-refractory mutation system-polymerase chain reaction (ARMS-PCR) method. Lipocalin-2 plasma protein expression has been measured by enzyme-linked immunosorbent assay (ELISA) technique, and compared with LCN2 plasma levels among the various types of cancer.
Methods
A case-control study has been conducted among 128 women aged between 18 years and 80 years from Jeddah city; 50% of whom were control and 50% were breast cancer patients. In the period between 2014 and 2016, blood samples of participants were taken from Medical Reference Clinics and King Abdulaziz University Hospital in Jeddah, Saudi Arabia. Participants filled their details regarding their family history. The tumors and history of psychological distress screened by mammography were included in the study. However, patients with severe complications were excluded.
Blood samples of participants were taken from Medical Reference Clinics and King Abdulaziz University Hospital in Jeddah. The ethical Committee (unit of biomedical ethics) approved the study at King Abdulaziz University (No.2//36/40591). Moreover, the study was conducted according to principles of Helsinki Declaration. Clinical data, Genomic DNA has been taken from whole blood and stored inside the Ethylenediaminetetraacetic acid (EDTA) coated tubes (Lavender top tube, Franklin Lakes, NJ, USA). For target, protein has taken from EDTA-plasma samples.
Genomic DNA was extracted by using ReliaPrepTM Blood gDNAMiniprep System kits. It was taken from whole blood and stored in the EDTA coated tubes (Lavender top tube, Franklin Lakes, NJ, USA). Lipocalin 2 gene amplification was carried out by PCR the used GoTaq® Green Master Mix. The first step in modeling the PCR system was the determination of Tm for the Tetra ARMS PCR primers. Touchdown PCR is a common method that employed a cycling program in which annealing temperature was adjusted. The starting annealing temperature was adjusted to several degrees, which was set at 60°C and 56°C. Additionally, the program employed had 4 major steps: Step 1: denaturation at 95°C for 2 min for one cycle. Step 2: 15 cycles for denaturation at 95°C for 45 seconds (s), annealing at 60°C for 45s and extension at 72°C for 1 min. Step 3: 20 cycles for denaturation at 95°C for 45s, extension at 72°C for 1 minute and annealing at 56°C for 45s. Step 4: Extension at 72°C for 5 minutes for one cycle. To confirm the PCR results, 3µl of PCR product was analyzed in 2% (w/v) agarose gel, the rest PCR product stored at a temperature of -2°C (Table 1).
Primer designing by primer blast software
Enzyme-linked immunosorbent assay using kit from Abcam’s Human LCN2 (NGAL) in vitro ELISA kit assessed plasma level of LCN2.
Statistical analysis
The Statistical Package of Social Sciences Version 20 (Armonk, NY: IBM Corp.) has been used. Data was presented as mean±standard deviation or number (percentage) wherever appropriate. The continuous variables were made between 2 groups using unpaired sample t-test and between more than 2 groups using one-way ANOVA (LSD) test. However, Chi-square was used for categorized data and the level of significance was p=0.05.
Results
Table 2 summarized the participants’ demographic characteristics. The age of the patients in this study was comparatively older than control. The early detection of tumors and positive family history were higher in patients (32.8%, 12.6%) as compared to control (23.4%, 10.9%) respectively.
Demographic characteristics of participant.
Table 3 showed the classifications based on the site, stage, and type of the tumor in all patients. The tumor stages consisted of some stages: stage 1 (54.7%), stage 3 (20.3%), stage 2 (18.8%) and stage 0 (6.2%) with the significant difference (p=0.0001). It has been observed that the tumor was dominant on the right side as compared to the left side (39.1%), then both sides (12.5%) with a considerable difference between them (p=0.001). The main tumor was realized to be invasive ductal carcinoma (73.4%) as compared to others (12.5%); ductal carcinoma in situ (10.9%); and invasive lobular carcinoma (3.1%), which also demonstrated a significant difference between them (p=0.0001). Expression of porgesterone receptor (PR) (73.4%), Estrogen Receptor (ER) (84.4%), and Human epidermal growth factor receptor 2 (HER2) (31.2%) receptors were positive expression, and the negative expression were PR: 26.6%, ER: 15.6% and HER2: 68.8%).
Description of the stage, site and type of the tumor of all patients.
Genotypes for LCN2 gene
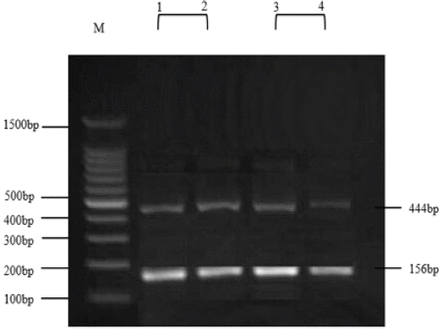
As mentioned in Figure 1, line 1-2 showed the genotyping results of LCN2 gene in the normal group. A double band of 444 and 156bp showed the presence of homozygous allele G that is the normal gene type. Line 3-4 showed the genotyping results of LCN2 gene in the patients group, which has also shown the presence of homozygous allele G and the normal gene type (double band of 444 and 156bp)
Genotypes for lipocalin-2 gene. Lane M: DNA marker. Lane 102: homozygous G/G normal gene type that produce double band of size 444 and 156bp (normal samples). Lane 3-4: homozygous G/G normal gene type that produce double band of size 444 and 156bp (breast cancer samples).
Lipocalin 2 plasma level and stages
Table 4 showed a considerably higher level of LCN2 plasma among patients as compared to controls (p=0.001). The comparison results of LCN2 plasma level with several types of breast cancer, indicated a considerably higher level in invasive lobular carcinoma (p=0.003) and invasive ductal carcinoma patients (p=0.001). No significant relationship was observed in comparing the LCN2 plasma level with different stages of breast cancer, stage I (p=0.676), II (p=0.780) and III (p=0.896).
Comparison of Lipocalin-2 plasma levels between different types of cancer and control.
Discussion
The present study has shown that there was no significant association between patients with breast cancer and LCN2 gene polymorphisms. Although, an accurate and efficient detection of rs11556770 polymorphism was made by tetra ARMS-PCR method with no special requirement of any equipment. In Kingdom of Saudi Arabia (KSA), breast cancer has been the most common cancer prevailed among women for past twelve years.19,20 Breast cancer is considered as the most common cancer among the females of Saudi Arabia despite of its low incidence. The factors increasing the risk of developing breast cancer include; genetic background or family history (15%), hormonal imbalance, increased breast density, and history of benign proliferative breast disease.20 Late stage occurrence in young women is a serious issue, particularly in countries like Saudi Arabia, where the development of breast cancer is rising with the time and evolving at younger ages.21 The cancer cells are predisposed due to constitutional inactivating mutations in genes encoding the protein that are important for DNA repairing.22 No research has examined the association between breast cancer and LCN2 polymorphism patients in Jeddah. Furthermore, a study conducted in Saudi Arabia indicated that type 1 diabetes patients, who had urinary LCN2 in their kidney tubules system, are more likely to have an ischemic injury.23 In addition, elevated LCN2 levels among patients can be used to forecast delayed graft function. Similarly, the existence of LCN2 among kidney donors can also be used for delayed graft function prediction.24 Hormonal changes including delayed parity, early menarch, and decreased breast feeding are common in the developing countries. These changes have a closer association with decreased exercise levels and increased intake of western diet. The impact of LCN2 gene polymorphisms (SNP rs11556770) on Breast cancer patients has been studied through the use of Tetra-Primer ARMS–PCR method. LCN2 plasma protein expression was measured by ELISA technique. Breast cancer patients with controls included 128 women from Jeddah city (64 breast cancer patients and 64 controls). The procedure includes Tetra ARMS-PCR; and this method has attained specificity and sensitivity. Thus, it can be concluded that this method is an efficient, as well as sufficient option for detection.25
Recently, LCN2 has meaningfully caught the attention of scientists and researchers as a modulator and as a biomarker of many varieties of human cancers. In particular, within breast cancer, it was realized that LCN2 levels were considerably higher in women breast cancer cells.7 In addition, the PCR analysis was employed for genotyping purposes. The LCN2 quality was implemented in the study to distinguish a SNP (rs11556770) (G/T) based on breast cancer database,26 in which a solitary nucleotide converts the amino acid glutamine (Gln) to histidine (His). The results indicated that 100% of genotype were (G/G) typical allele for the populace of breast cancer, residing in Jeddah. No relationship has been recognized between rs11556770 SNP and risk of breast cancer.
In addition, the research also aimed to determine LCN2 plasma protein expression through ELISA, amongst control and distinctive sorts of breast cancer. The results realized that LCN2 plasma levels were higher in patients as compared to controls (p=0.001) and was significantly elevated among patients with invasive lobular carcinoma (p=0.001) and invasive ductal carcinoma (p=0.003). In contrast, other study found a positive relationship between swelling grade and LCN2 levels.27 It was also noted that breast cancer cells contained high LCN2 levels and elevated LCN2 expression. It connected the findings to expectation of breast cancer incidence, expanded capacity for carcinoma increase, pitiable histological reviewing, and lymph node metastasis.28 Usually, there are significant LCN2 traces in tumor stroma in the stage II and stage III of breast cancer. It contrasted the study results, which showed that there was no significance level of LCN2 plasma in the breast cancer stages I, II and III (p=0.676, p=0.780, p=0.896).
In conclusion, the analysis showed that there was no significant relationship between the breast cancer patients and LCN2 gene polymorphisms. However, tetra ARMS-PCR method was an efficient and accurate approach for the detection of rs11556770 polymorphism without need of any special equipment. On the contrary, LCN2 protein expression in plasma level was considerably elevated among patients as compared to controls, particularly among patients demonstrating invasive ductal carcinoma. The small sample size and concerning a small geographical area is a major limitation of the study. Moreover, we did not investigate on LCN2 from a clinical perspective to apply in diagnostic procedures. For further study, it has been recommended to conduct an expanded sample of breast cancer patients and also on LCN2 from a clinical perspective to apply it for diagnostic purposes.
Acknowledgment
Authors are very thankful to all the associated personnel in any reference that contributed in/for the purpose of this study.
Footnotes
Disclosure. Authors have no conflict of interests, and the work was not supported or funded by any drug company.
- Received March 25, 2018.
- Accepted May 16, 2018.
- Copyright: © Saudi Medical Journal
This is an open-access article distributed under the terms of the Creative Commons Attribution-Noncommercial-Share Alike 3.0 Unported, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.