Abstract
Objectives: To determine the distribution and resistance profiles of Gram-negative bacteria (GNB) in intensive care units (ICUs) at King Abdullah Hospital in Bisha, Saudi Arabia.
Methods: A record based retrospective study was conducted from December 2016 to January 2018. In total, 3736 non-duplicate clinical specimens from the general intensive care unit (ICU), neonatal ICU (NICU), and coronary CU (CCU) were analyzed for pathogens.
Results: Of 3736 specimens, 9.6% (358) were positive for pathogens, and GNB constituted the majority (290/358; 81%). Acinetobacter is predominant in the general ICU, whereas Klebsiella pneumoniae is common in the NICU and CCU. Overall, GNB revealed a high resistance rate for cefuroxime (75.8%) trimethoprim/sulfamethoxazole (73.4%), cefotaxime (72.9%), aztreonam (64.6%), piperacillin (62.1%), and ciprofloxacin (61.5%). Acinetobacter revealed a high resistance (93.4% to 97.5%) to all antimicrobials except colistin (4%). Klebsiella pneumoniae showed a high resistance to trimethoprim/sulfamethoxazole (71.8%), cefotaxime (71.4%) and aztreonam (65.2%). Pseudomonas aeruginosa showed good activity for aminoglycosides but increasing resistance for cephalosporins and meropenem. GNB exhibited a high rate of multi-drug-resistant (MDR) phenotypes (67.9%) with a higher level among Acinetobacter spp. (97.5%). There were no significant differences in the resistance rates of GNB from different ICUs except for imipenem (p=0.002) and ciprofloxacin (p=0.003).
Conclusions: Increased antimicrobial resistance with high proportions of MDR patterns were found among GNB from ICUs. Comprehensive surveillance programs are needed to track the origins and emergence pathways of resistant pathogens.
In the last decade, antimicrobial-resistant pathogens have been continuing to spread as causative agents of nosocomial infections in intensive care units (ICUs).1,2 Gram-negative bacteria (GNB) such as Acinetobacter, Pseudomonas aeruginosa, Klebsiella spp. and Escherichia coli are responsible for ICU infections.1 They are a common cause of infections of the primary bloodstream, pulmonary tract, urinary tract, postsurgical sites, skin, and soft tissue.3 Patients in the ICU are at higher risk of acquiring nosocomial infections caused by antimicrobial-resistant pathogens.4,5 Infections caused by multidrug-resistant (MDR) bacteria have become the main factor of increasing morbidity and mortality among patients in ICUs, which makes them a great health challenge for hospital authorities.4,6 The mortality rate found to be increased more than twice among infected ICU patients comparable to non-infected patients.1 One study found that the risk of dying among ICUs neonates having nosocomial infections was greater more than 3 times compared to those who without the illness.7
The rates of MDR patterns among Gram-negative pathogens in ICUs are several times higher than in general hospital settings.1 The dissemination of such pathogens is emerging as a global health problem.8 Worldwide surveillance studies have been conducted to assess bacterial pathogens and their resistance profile in ICUs.9,10 In Saudi Arabia, several local studies in tertiary hospitals have tracked the resistance rates of pathogens in ICU settings.11-13 However, such data are scarce in certain areas, including Bisha Province in the southwest of the country. The availability of regional data on the resistance rates is fundamental to implementing effective treatment protocols against infectious agents and might help to prevent infections with MDR pathogens at the local level.9,14
The appropriate identification of ICU pathogens and their MDR phenotypes plays a crucial role in the treatment process and in infection control measures.9,15 This is the first study with the aim of determining the distribution of GNB and their resistance rates collected from clinical samples of patients in ICUs at a tertiary care hospital in Bisha, Saudi Arabia. We also aimed to determine the MDR patterns of the isolates associated with patients and different ICUs.
Methods
A record based retrospective study was conducted over one year from December 2016 to January 2018 at King Abdullah Hospital, Bisha Province, Southwest Saudi Arabia. This hospital is a 365-bed tertiary care center that offers different specialized services. It provides referral healthcare services to an estimated 17,162 inpatients and 215,322 outpatients per year for patients living in the province, suburbs, and neighboring provinces. The hospital has 3 different ICUs containing 43 beds, including a general ICU (16 beds), neonatal ICU (NICU; 23 beds), and coronary CU (CCU; 4 beds). These units are supported by a multi-disciplinary care team. Ethical approval for this study was obtained from the Research and Ethical Committee of the College of Medicine at the University of Bisha.
A standard data-collection form was used to collect patients’ general information, the type of ICU, isolated pathogens, the type of specimens, and antimicrobial susceptibility profiles from the laboratory information system. A total of 3736 non-duplicate clinical specimens of patients (n=3736) at 3 ICUs were retrieved and analyzed, including blood (n=1565), urine (n=1089), sputum (n=869), tracheal aspirate (n=95), wound swab (n=67), eye swab (n=24), throat swab (n=12), umbilical discharge (n=11), ear swab (n=3), and high vaginal swab data (n=1). The positive microbiological cultures of these specimens were retrieved and analyzed. Only patient specimens with complete information were recorded and included in the data analysis. Additional isolates from the same specimens were excluded.
Isolation and identification of pathogens
The hospital microbiology laboratory received the clinical specimens and processed them according to the standard operating procedures for the isolation and identification of possible pathogens.16 Depending on the origin of the samples, each clinical specimen was cultured on blood agar, (Oxoid, Basingstoke, United Kingdom) MacConkey agar (Oxoid, Basingstoke, United Kingdom), chocolate blood agar (Oxoid, Basingstoke, United Kingdom), or brain heart infusion broth (Oxoid, Basingstoke, United Kingdom). They were then incubated aerobically at 37°C for 24 to 48 hours except for the blood culture, which was incubated for 5 to 7 days.
Preliminary identification of some isolates was performed based on the colonial morphology, Gram stain, and routine rapid biochemical tests such as catalase, indole, and oxidase tests. Full identification of the isolate was then performed using the Phoenix system identification method (Becton Dickinson, Sparks, Maryland, USA). The Phoenix panels were inoculated as per the manufacturer’s guidelines. Depending on the site of infections and the types of specimens, the significant growth of each pathogen was identified and processed for antimicrobial susceptibility testing.
Antimicrobial susceptibility testing
The susceptibility test of GNB was carried out according to the recommendations of the Clinical and Laboratory Standards Institute (CLSI).17-19 Identified strains were tested in vitro against several classes of antimicrobial drugs using the Phoenix automated microbiology system.20 The following antimicrobial agents were examined: amikacin (30 µg), aztreonam (30 µg), cefepime (30 µg), cefotaxime (30 µg), ceftazidime (30 µg), cefuroxime (30 µg), ciprofloxacin (5 µg), colistin (10 µg), gentamicin (10 µg), imipenem (10 µg), meropenem (10 µg), piperacillin (100 µg), piperacillin/tazobactam (110 µg), tobramycin (10 µg), and trimethoprim/sulfamethoxazole (23.75 µg/1.25 µg). Quality control and maintenance were achieved according to the manufacturer’s guidelines. Multidrug-resistant isolates were identified based on resistance to at least 3 different antimicrobial classes as described previously.21
Statistical analysis
The Statistical Package for Social Sciences (SPSS version 22)(Armonk, NY: IBM Corp.) was used for data entry and analysis. The distribution of bacterial isolates and their resistance patterns across different ICUs, age groups, and genders were expressed as proportions. Susceptibility test results were classified as “resistant” or “susceptible.” Missing data were excluded from the analysis. The MDR patterns of the isolates were compared between ICUs and across 5 patient age categories: infants (>0.0 days to 1 year), children (>1 to 18 years), young adults (>19 to 39 years), adults (40 to 64 years), and the elderly (>65 years). A Chi-squared test was used to compare every 2 variables with a p-value less than or equal 0.05 considered as statistically significant.
Results
Bacterial isolates
Of the 3736 non-duplicate specimens cultured for possible pathogens, 9.6% (358) yielded significant growth for various microorganisms. Wound swabs yielded the highest culture positivity (n=21/67; 31.3%), followed by sputum (n=188/869; 21.6%), tracheal aspirates (n=19/95; 20%), umbilical swabs (n=1/11; 9.1%) throat and eye swabs (n=1/11; 8.3% for each), urine (n=30/1089; 2.8%) and blood (n=28/1565; 1.8%).
Gram-negative bacteria constituted 81% (290/358) of the isolates, while 15.9% (57/358) were Gram-positive cocci, and 3.1% (11/358) were yeast cells. The majority of GNB was recovered from male patients (n=173; 59.7%). Elderly patients (n=128; 44.1%) were the most commonly infected group, followed by young adults (n=85; 29.3%), adults (n=37; 12.8%), infants (n=30; 10.3%), and children (n=10; 3.4%).
Table 1 summarizes the distribution of GNB (n=290) recovered from various clinical specimens of ICU patients. Most of the isolates were recovered from samples of sputum (n=188), urine (n=30), blood (n=28), and wound sites (n=21). Acinetobacter spp. were the predominant isolates (n=79; 27.2%), followed by P. aeruginosa (n=69; 23.8%) and K. pneumoniae (n=54; 18.6%).
Distribution of 290 Gram-negative isolates recovered from various clinical samples of patients at intensive care units.
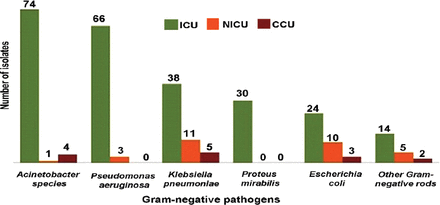
Figure 1 shows the distribution of isolates according to the type of ICU. In the general ICU, Acinetobacter spp. were the predominant pathogens, followed by P. aeruginosa and K. pneumoniae. Klebsiella pneumoniae was the most frequent isolate from the NICU and CCU.
Frequency of Gram-negative pathogens among 3 intensive care units (general ICU, neonatal ICU and coronary care unit. Others Gram-negative rods include: Morganella morganii (n=7), Citrobacter freundii (n=1), Providencia stuartii (n=4), Enterobacter cloacae (n=3), Stenotrophomonas maltophilia (n=2), Unidentified Gram-negative bacilli (n=4).
Antimicrobial susceptibility of the isolates
Table 2 summarizes susceptibility of GNB to several types of antimicrobial drugs. Overall, the highest resistant rate was found for trimethoprim/sulfamethoxazole (79%) followed by cefotaxime (72.9%), cefepime (70%) and aztreonam (64.6%). The lowest resistance rate was reported for colistin (7.5%).
Proportions of antimicrobial resistance of Gram-negative isolates (n=290) recovered from patients at intensive care units at King Abdullah Hospital, Bisha province, Saudi Arabia.
Of all the isolates, E. coli had the lowest resistant rates to the tested antimicrobials. Acinetobacter spp. revealed high resistance rates ranging from 93.4% to 97.5% for all tested antimicrobial agents except for colistin. Among K. pneumoniae, the rates of resistance were counted 71.8% for trimethoprim/sulfamethoxazole 71.4% for cefuroxime and cefotaxime; 67.3% for cefepime, 65.2% for aztreonam and 64.2% for piperacillin. Pseudomonas aeruginosa showed low to moderate resistance rates for aminoglycosides (18.8% for amikacin; 20% for tobramycin, 31.7% for gentamicin). But, increasing resistance rates were observed to cephalosporins, aztreonam, meropenem, piperacillin, and piperacillin/tazobactam.
Proteus mirabilis showed the highest resistance rates for trimethoprim/sulfamethoxazole (87%) and cefuroxime (58.6%). Other GNB such as Morganella morganii, Citrobacter freundii, Providencia stuartii, Enterobacter cloacae, Stenotrophomonas maltophilia, and unidentified GNB revealed the highest resistance rates for cephalosporins (80% to 95%) and ciprofloxacin (72.2%).
Table 3 summarizes the antimicrobial susceptibility rates of the isolates recovered from the 3 ICUs. There were no significant differences in resistance rates of the isolates collected from the 3 ICUs except for imipenem (p=0.002) and ciprofloxacin (p=0.003). Gram-negative isolates from the NICU were more susceptible to imipenem (75.9%) and ciprofloxacin (71.8%).
Comparison of antimicrobial resistance rates of Gram-negative pathogens collected from patients at three intensive care units, King Abdullah Hospital, Bisha, Saudi Arabia.
Multidrug-resistance phenotype
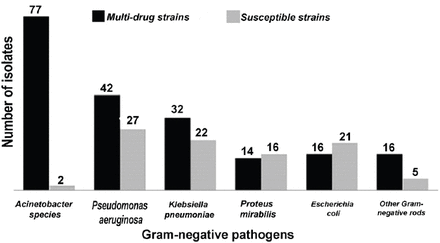
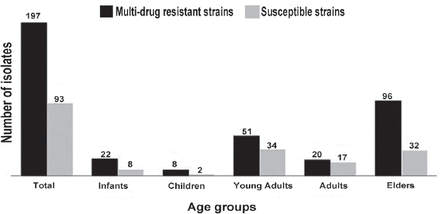
Of a total of 290 Gram-negative isolates, 67.9% exhibited the MDR phenotype. Acinetobacter spp. yielded the highest rates of MDR patterns (n=77/79; 97.5%) (Figure 2). Multi-drug-resistant patterns differed significantly according to the patient age (p=0.045). Isolates from elderly patients (age more than 60 years old) were more resistant to antimicrobial drugs when compared to isolates from other age groups (Figure 3). There was no statistically significant association (p=0.563) between the presence of MDR isolates and type of ICU.
Frequency of multidrug-resistant (MDR) Gram-negative bacteria (n=290) recovered from patients of all age groups at intensive care units. Others Gram-negative rods include: Morganella morganii (n=7), Citrobacter freundii (n=1), Providencia stuartii (n=4), Enterobacter cloacae (n=3), Stenotrophomonas maltophilia (n=2), Unidentified Gram-negative bacilli (n=4).
Frequency of multidrug-resistant (MDR) Gram-negative bacteria (n=290) recovered from patients of all age groups at intensive care units.
Discussion
Intensive care units are becoming main reservoirs of resistant pathogens that constitute a challenge for clinical practice and hospital authorities.2 Thus far, there has been no published information on antimicrobial resistance patterns for GNB in ICUs at King Abdullah Hospital, Bisha Province, Saudi Arabia.
In the present study, the majority of bacterial pathogens were recovered from male patients. The possible explanation might be that ICUs in this hospital is predominantly populated with infected male patients than female patients. Moreover, most of the isolates were recovered from the respiratory system. This result agrees with a previous study in Riyadh capital, where the majority of bacterial isolates recovered from ICU patients were also from the respiratory tract.22 Surveillance studies in ICUs also indicated that the majority of bacterial isolates were from the respiratory tract.5,9 Our findings might indicate that respiratory infections are the most common in our ICUs.
The most commonly isolated pathogens in this study were Acinetobacter spp. (27.2%) followed by P. aeruginosa (23.8%) and K. pneumoniae (18.6%). These figures could be compared with those reported in a retrospective cohort study conducted at Riyadh Military Hospital, where A. baumannii represented 40.9% of all ICU isolates, followed by K. pneumoniae (19.4%) and P. aeruginosa (16.3%).22 Consistently, such GNB isolates were the most frequent pathogens in 6 years of epidemiologic surveillance in Saudi Arabia for ventilator-associated pneumonia at a tertiary-care ICU.23 The predominance of GNB in ICUs was also reported in surveillance programs conducted in Europe and the USA.10,24 The spread of these etiological agents might be linked to contaminated respiratory equipment and transmission via the hands of healthcare workers in ICU settings. Therefore, the transmission of GNB in ICUs could be prevented through reinforcing appropriate control measures coupled with examination of the rate of bacterial contamination of the hands of healthcare workers and the ICU environment.25
In the present study, Acinetobacter spp. demonstrated high resistance rates for carbapenems such as imipenem (97.5%) and meropenem (96.2%). The high resistant rates pose a major concern in our ICU as carbapenems are the drug of choice for Acinetobacter infections. Our findings suggest the existence of carbapenemase-producing strains, which has also been reported in other studies in Saudi Arabia and Gulf Cooperation Council States.26,27 However, determining the carbapenem-resistance mechanisms could lead to improvements in the outcomes of infections caused by these bacteria.26 Heavy uses of antimicrobial agents and invasive procedures have been found to be risk factors for the acquisition of antimicrobial-resistant pathogens in ICUs.5,28,29 The role of healthcare workers in the transmission of environmental Acinetobacter clones within the ICU has been reported in a previous study in Saudi Arabia.30
Our results showed that more than 60% of K. pneumoniae isolates were resistant to cephalosporins, trimethoprim/sulfamethoxazole, aztreonam, piperacillin, and piperacillin/tazobactam. The resistance rates for aminoglycosides, carbapenems, and ciprofloxacin remained between 40 and 55%. In another setting in Saudi Arabia, K. pneumoniae demonstrated the highest resistance rates for ceftriaxone (59.4%), aztreonam (58.3%), and ceftazidime (58.3%). Higher usage of broad-spectrum cephalosporins increases the prevalence of resistant bacteria in ICU.31 The extensive use of these agents also promotes the emergence of extended-spectrum beta-lactamase (ESBL) and AmpC beta-lactamase producing isolates.32 The dissemination of K. pneumoniae and E. coli isolates harboring diverse resistant beta-lactamase resistance genes has been studied previously, which revealed resistance to cephalosporins and other antimicrobial classes.33,34
In the present study, P. aeruginosa demonstrated good susceptibility to amikacin (81.2%), gentamicin (68.3%), and imipenem (61.8%). In the USA and Europe, amikacin exhibited good activity against P. aeruginosa strains from ICU patients (97.3% in the USA and 84.9% in EU).10 Furthermore, Radji et al found that amikacin was the most effective (84.4%) antibiotic against P. aeruginosa isolated from patients at ICU in a hospital in Indonesia, followed by imipenem (81.2%), and meropenem (75.0%).1
The presence of diverse resistant pathogens is an emerging clinical problem in ICUs, including neonatal, pediatric, and various adult critical care units.9 In the comparison between the isolates from the 3 ICUs, we found that Acinetobacter spp. are the most frequent isolates from the adult ICU, whereas K. pneumoniae is the predominant isolate from the CCU and NICU. In contrast, reports from African countries and India found that Staphylococcus spp. were the predominant isolates from patients in the pediatric ICU and NICU.29,35,36 The spread of such diverse pathogens could be explained by horizontal transmission from colonized visitors or healthcare workers to the pediatric population in the ICU.36
As in previous studies,29,35 our findings revealed high resistance rates among isolates from the 3 different ICUs. However, significantly higher resistance rates were recorded among isolates from the general ICU than the NICU for imipenem and ciprofloxacin. A possible explanation might be the extensive use of these drugs in the general ICU. However, fluoroquinolones are not used in our pediatric ICU. One study indicated that carbapenems such as imipenem remain a reliable option for the treatment of severe infections caused by GNB in the pediatric population.37
The proportion of MDR among GNB was high (67.9%), and this finding agrees with studies from an eastern province of Saudi Arabia38 and elsewhere.8,9 The MDR rates are questionable, particularly for Acinetobacter spp. (97.5%), which makes ICU infections caused by GNB difficult to treat in our setting. A study in the similar southern region of the country at Aseer Central Hospital, Abha, found that non-compliance with hand hygiene among healthcare workers spread MDR strains from patient to patient in ICUs.39 Furthermore, cross infections among inpatients, ICU procedures, and patients with chronic disease have led to the emergence of MDR strains in ICUs.9,22 Prolonged hospital ICU admission coupled with unnecessary antibiotic administration might also increase the spread of MDR pathogens. Therefore, the common risk factors that could be associated with escalating MDR patterns among ICU pathogens should be defined in our hospital. In a recent study in Saudi Arabia,38 elderly patients tended to acquire more MDR isolates than other age groups. A weakened immune system associated with chronic illness among older patients was found to be a risk factor for the acquisition of MDR pathogens.28
Study limitations
The present study has several limitations need to be addressed in the future researches. Firstly, some clinical data were missed or incomplete, leading to exclude several clinical specimens from the study. Secondly, identifications of MDR isolates were based on the limited number of antimicrobial classes that prescribed in the hospital. Thirdly, the number of clinical isolates from CCU and NICU were small compared to general ICU isolates which leading to restrict the statistical analysis. Fourthly, the frequency of ESBL producing GNB was not reported which need further investigations to determine such resistant mechanisms.
In conclusion, this study revealed that Acinetobacter spp., P. aeruginosa, and K. pneumoniae are the most common GNB associated with ICU infections in this tertiary hospital. Isolates from different ICUs showed high resistance rates to most antimicrobial agents, and most of them (67.9%) exhibited MDR patterns, with the highest frequency occurring among Acinetobacter spp. (97.7%). The high rates of antimicrobial resistance are a critical condition that calls for comprehensive surveillance programs to track the origins and emergence pathways of resistant pathogens. Developing local antimicrobial stewardship programs and continuous monitoring of antimicrobial susceptibility might be useful to preclude inappropriate antimicrobial use and the emergence of antimicrobial resistance patterns.
Footnotes
Disclosure. Authors have no conflict of interests, and the work was not supported or funded by any drug company.
- Received May 16, 2018.
- Accepted August 22, 2018.
- Copyright: © Saudi Medical Journal
This is an open-access article distributed under the terms of the Creative Commons Attribution-Noncommercial-Share Alike 3.0 Unported, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.