Abstract
The detection of carbapenem-resistant organisms (CROs) represents a substantial challenge for many clinical laboratories. In this review, several phenotypic and non-phenotypic methods for detecting CROs are discussed. However, no consensus has yet been reached with regards to the single most optimal method. Due to differences in carbapenem-resistant activity between carbapenemases, the simultaneous use of 2 or more phenotypic detection methods can improve the detection of CROs compared with a single technique. Molecular methods are currently favored because the majority can be performed rapidly with a high level of accuracy. Whole-genome sequencing (WGS) yields unambiguous data pertaining to complete analysis of the entire genome and may ultimately become a highly powerful tool in routine clinical settings. However, WGS is still relatively expensive and requires an automated data interpretation system. The routine implementation of this technique in clinical laboratories may not occur for several years, particularly in developing countries.
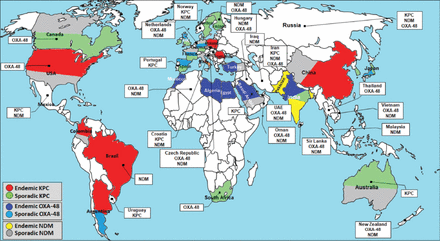
Within the last decade, antimicrobial resistance in gram-negative bacilli has become a major concern in healthcare settings worldwide.1 Gram-negative bacilli that produce extended-spectrum β-lactamases (ESBLs) have the ability to resist several antibacterial agents including third generation cephalosporins and monobactams. These bacterial species include members of the Enterobacteriaceae family, Acinetobacter baumannii and Pseudomonas aeruginosa. Among available β-lactam antibiotics, carbapenems are capable to resist many β-lactamase enzymes.2 This feature renders them the last resort drugs to treat serious infections of many ESBL-producing gram-negative bacilli,3,4 However, overuse of carbapenems has led to the emergence of carbapenem-resistant Enterobacteriaceae (CRE), carbapenem-resistant A. baumannii (CRAB) and carbapenem-resistant P. aeruginosa (CRP). The carbapenem-resistant mechanisms vary among CRE, CRP and CRAB. For example, CRE isolates are capable of inactivating carbapenem via the production of carbapenemase enzymes; CRP and CRAB can resist carbapenems via a combination of carbapenemase enzymes, mutation of porins and efflux pump overexpression.5 There are many carbapenemases that have been identified and categorized into classes. It is worth noting that the Ambler molecular classes A carbapenemases (KPC), B metallo-β-lactamases (MBLs) (VIM, IMP, NDM), and D oxacillinases (OXA-48) are considered the most clinically important carbapenemases.4,6 These classes are now commonly found in CRE (for example, K. pneumoniae) (Figure 1); OXA-51-like, OXA-23, OXA-24/40, OXA-58, OXA-143, and OXA-235-like enzymes are associated with CRAB isolates that have been associated with serious nosocomial infections in various countries.4,7-14 New Delhi MBL (NDM-1), for example, was first described in India and has been recently found in Asia, North America, Europe and Australia.13 Likewise, oxacillinase-48-type (OXA-48) enzyme was first reported in K. pneumoniae in Turkey and has recently spread across Europe and the Middle East.10 Although Klebsiella pneumoniae carbapenemase (KPC) enzyme was first identified in K. pneumoniae isolates in North Carolina in the United States, in 1996,15,16 KPC producers have now spread across 38 American states. Klebsiella pneumoniae carbapenemase (KPC) has been recently reported in Canada, South America, Europe, Australia, China and India (Figure 1). Nowadays, the global spread of CRE is of significant concern to the international health community because only limited antimicrobial alternatives are available to confront carbapenem-resistant gram-negative bacilli (CR-GNB) infections. Therefore, the early detection of CR-GNB, proper hand washing practices, and the prudent use of antibiotics are considered to be the most effective control methods for CR-GNB infections. The accurate detection of carbapenem-resistant organisms (CROs) is a challenge for many clinical laboratories for both clinical and screening cultures.17 Although routine antibiotic susceptibility tests (ASTs) are still required to select the most appropriate treatment option, differences in carbapenem-resistant activity between carbapenemases render the detection of carbapenem-resistance using only ASTs more complicated. For example, Ambler class A (KPC) hydrolyze nearly all β-lactams, but their hydrolytic activity can be inhibited by some β-lactamase inhibitors such as boric acid, clavulanic acid and tazobactam. However, Ambler class B or MBLs (NDM, VIM, IMP) have a broad range of activity against penicillins, cephalosporins and carbapenems; Ambler class D (OXA-48) types possess poor activity against cephalosporins and carbapenems. Furthermore, the activity of Ambler class D is not inhibited by current available β-lactamase inhibitors.18,19 Many phenotypic and genotypic (molecular) detection methods can identify carbapenem-resistant pathogens and discriminate between carbapenemase classes. However, they vary in their sensitivity, performance requirements and cost. The aim of this review is to provide an overview of all existing methods for the detection of CR-GNB.
Worldwide distribution of the most clinically important carbapenemase genes (Klebsiella pneumoniae carbapenemase (KPC), New Delhi Metallo-β-lactamase (NDM), and oxacillinase-48 (OXA-48). Carbapenemases listed in white rectangles have been recently recorded.
Detection methods
In the last 10 years, several methods for detecting CR-GNB have become available. These methods include phenotypic-based methods that detect the activity of carbapenemase enzymes such as growth-based assays, rapid colorimetric methods, immunochromatographic (IC) assays, and molecular-based methods (e.g., polymerase chain reaction (PCR)-based methods and whole-genome sequencing (WGS)).
Phenotypic-based methods. Antibiotic susceptibility test
Susceptibility pattern by disc diffusion is considered to be a first-line method for early detection of carbapenemases; ertapenem has been described as the most sensitive indicator for the activity of carbapenemases.20 Minimum inhibitory concentration (MIC) breakpoints can also be used as a confirmatory testing method, and this test can be carried out using different platforms including the E-test, broth microdilutions, agar dilutions and automated antimicrobial susceptibility testing (AST) systems (e.g., Vitek 2, MicroScan, Phoenix). Such testing methods have demonstrated variability in their ability to detect carbapenem-resistance. Although the determination of carbapenem MIC breakpoints suggested by the European Committee on Antimicrobial Susceptibility Testing (EUCAST) is sometimes useful for predicting CP-GNB, some CP-GNB isolates remain susceptible or intermediate to many carbapenems. For example, some Enterobacteriaceae isolates exhibit susceptibility to ertapenem, meropenem and imipenem but carry the blaKPC gene.21,22 In 2009, the Clinical and Laboratory Standards Institute (CLSI) issued recommendations for the identification of carbapenemase-producing organisms (CROs) based on the MIC breakpoints of 2 mg/L or zone diameters <22mm for all carbapenems.23 One year later, the CLSI also recommended that Enterobacteriaceae isolates that are resistant to all third-generation cephalosporins and exhibit carbapenem MIC breakpoints of 2 or 4 µg/ml should be confirmed by the modified Hodge test (MHT).24 This approach demonstrates a good sensitivity for the detection of KPC but a low sensitivity to NDM and IMP. The CLSI guideline are also plagued by another problem some CRE producing OXA-48-type enzymes may be sensitive to third-generation cephalosporins.23,24 In 2016, the CLSI suggested that confirmatory tests such as the MHT, the Carba NP test and/or a molecular technique should be performed when Enterobacteriaceae isolates are associated with a suspicion of carbepenemase production based on updated carbapenem breakpoints (imipenem or meropenem or ertapenem MICs of at least ≥2µg/mL) (Table 1).25,26 Although both CLSI and EUCAST have attempted to improve the interpretation of routine ASTs to detect CROs based on carbapenem breakpoints, these methods still fail to detect all or most of CROs.27 Indeed, routine ASTs are necessary and helpful for treatment purposes but not for screening and epidemiological purposes.
Clinical breakpoints for carbapenems for Enterobacteriaceae, Pseudomonas, and Acinetobacter according to CLSI 2018 and EUCAST 2018 guidelines.
Multi-disk mechanism testing and combined disk synergy tests
Multi-disk diffusion tests are phenotypic methods that can distinguish between some types of carbapenemase. These methods are based on the synergy between β-lactamase inhibitors and carbapenems. These inhibitors include boronic acids for KPC detection, dipicolinic acid or Ethylenediaminetetraacetic acid (EDTA) for MBL, clavulanate (an inhibitor of ESBL), cloxacillin (an inhibitor of AmpC) and avibactam (NXL104) for OXA-48.28-30 Multi-disk tests have been widely used due to their simplicity and low cost. These tests vary in their sensitivity, which ranges from 90% to 100%.28,31 Several commercialization phenotypic multi-disk tests have been developed to identify and differentiate different types of carbapenemase. Mastdiscs ID inhibitor combination detection disks (MDI), for example, is a new method that was developed by Mast Diagnostics to identify carbapenemases based on a simple calculation of the inhibition zone of combined disks, incorporating specific enzyme inhibitors. In one study, this method exhibited good discriminatory power in KPC and MBL detection among K. pneumoniae, but it was not able to distinguish between OXA-48-types genes and different MBL genes such as NDM, VIM and IMP.12 The Rosco Diagnostica Neo-Sensitabs (RDS) is also another carbapenemase detection test, this method was evaluated by Doyle et al.32 These authors concluded that both phenotypic tests (MDI and RDS) lack sensitivity for detecting OXA-48-like enzymes.
Recently, a new phenotypic test, the OXA-48 disk test, was introduced for differentiating OXA-48-producing CRE from those producing other carbapenemases. This test has been shown to exhibit excellent performance for detecting OXA-48 (sensitivity: 96.3%).33 In general, multi-disc tests are simple to perform and interpret, and they appear to be useful for clinical laboratories that do not have access to or cannot afford molecular techniques.
Chromogenic-based media for screening purposes
Carbapenem-resistant Enterobacteriaceae carriers typically represent the primary sources of CRE spread in healthcare settings, and all high-risk patients (e.g., elderly patients, patients in intensive care units (ICUs), immunocompromised patients and patients arriving from CRE-epidemic areas) should be screened. To detect CRE gastrointestinal carriers, fecal samples or rectal/perirectal swabs are recommended.34 Chromogenic-based media are considered to be among the optimal screening methods for rapid detection of CRE.35 They are based on a chromogenic enzyme substrate and specific antibiotics that render them selective for a specific resistance feature.27 There are many commercial chromogenic agars that are supplemented with carbapenems and are currently used for detecting carbapenemase-producing bacteria: CHROMagar KPC (CHROMagar, France), chromID OXA-48, chrom ID Carba, Colorex KPC (Biomed Diagnostics, USA), Hardy CHROM (Hardy Diagnostics, USA), Ramba Chrom-KPC (Gibson Bioscience, USA), and Brilliance CRE (Thermo Diagnostics, USA). Many previous studies have evaluated most of these media in terms of their ability to detect specific carbapenemase classes, these media exhibited varied sensitivities that ranged between 13 and 100%.24,27,36-38 A low sensitivity of detecting OXA-48 producers was commonly observed in some media. Nordmann et al39 developed a novel screening medium (a nonchromogenic medium) called Supercarba that exhibited a high sensitivity (96.5%) for all carbapenemase including OXA-48 producers.39,40 However, this medium appears to detect only carbapenemases in lactose-fermenting bacteria.27 Its limited shelf life (7-10 days), once prepared, is also another disadvantage in clinical microbiology laboratories.40,41 The Supercarba medium has been adapted to a chromogenic form called mSuperCarba. This medium exhibited 100% sensitivity and specificity for detecting KPC, MBL and OXA-48-type-producing isolates of CPE. Also, the shelf life of mSuperCarba medium has been improved to up to one month.41,42 Indeed, chromogenic-based media as a screening method are helpful and can decrease the turnaround time for the detection of carbapenemase producers by excluding the enrichment step that can require 24 h.24
MHT
The MHT is a simple phenotypic test for the detecting of carbapenemase enzymes among Enterobacteriaceae isolates. This test depends on the ability of a carbapenemase-producing strain to decrease the carbapenem concentration and enable the growth of a carbapenem-susceptible Escherichia coli strain.43 The MHT was recommended by the CLSI from 2009-2017 as a confirmatory test for carbapenemases based on its capability to detect KPC producers, and it is therefore used in many clinical microbiology laboratories. It also has good sensitivity in term of detecting other types of carbapenemases including VIM, IMP and OXA-48.26,30 However, the MHT is characterized by low sensitivity to other carbapenemases such as NDM, MBL, some OXA types, and SME.43 Furthermore, the MHT suffers from a lack of specificity and may yield false-positive results when detecting some AmpC-producing isolates combined with porin mutations.30 Therefore, the MHT was omitted in the current edition of the CLSI 2018 due to the issue of false positives and false-negative results.25 Despite these limitations, the MHT remains useful for preliminary screening of large numbers of CRE isolates in many clinical laboratories due to its simplicity, cost-effectiveness and moderate sensitivity.30
Carba NP
The Carba NP is a phenotypic colorimetric assay that is used to detect CPE. The Carba NP was developed in 2012 by Patrice Nordmann, Laurent Poirel and Laurent Dortet.18 There is one commercially available Carba NP tests (marketed as the RAPIDEC CARBA NP), and it relies on measuring imipenem hydrolysis that can change pH and result in a color change of a colorimetric pH indicator (phenol red).30 This technique has many advantages over other phenotypic tests: simplicity, speed, cost-effectiveness, sensitivity and specificity.44 The Carba NP has been recommended by the CLSI and EUCAST as a confirmatory test for carbapenemase producers in Enterobacteriaceae and other CR-GNB.25,43,44 In many studies, the sensitivity of the Carba NP for most Carbapenemases ranged from 73-100%.18,30,45,46 However, the Carba NP exhibits low sensitivity to some carbapenemases that have low imipenem hydrolysis activity such as OXA-48 and some class A carbapenemases such as GES-5 and SME-1.21,46 Another study reported that the Carba NP exhibited equivocal results for isolates cultured on MacConkey and Drigalski agar. This study suggested that both media have a negative effect on the Carba NP.47
Carbapenem inactivation method
The carbapenem inactivation method (CIM) is a novel phenotypic method that was first introduced by van der Zwaluw and co-workers in 2015.48 The CIM relies on the in-vitro detection of carbapenem inactivation by CPOs. This method involves suspending bacterial colonies in water containing carbapenem disks (usually a 10µg meropenem) and incubating the set-up for 2 h. The meropenem disk is then positioned onto Mueller-Hinton agar (MHA) inoculated with a carbapenem-sensitive E. coli strain. The absence of a large zone of inhibition denotes that a meropenem disk has lost its antimicrobial activity as a result of producing carbapenemase enzyme by the organism of interest. If the meropenem disk is still active, a clear inhabitation zone can be observed around the disk that indicates the organism of interest is not a carbapenemase-producer. The CIM has been described as a straightforward, cost-effective and highly sensitive method with excellent specificity that is available to many clinical laboratories.48 It is also comparable in cost to the MHT, but it is more sensitive and specific and is currently recommended by CLSI 2018 in the form of the modified CIM (mCIM).25,43,48 Other comparison studies have shown that the CIM is more accurate than the Carba NP and MHT at detecting carbapenem producers among Enterobacteriaceae. These studies have also reported that the CIM exhibited equal and/or better sensitivity at detecting OXA-48.49-51 Although the CIM requires more time (24 h) to cultivate the organism of interest, it is simple, cheap, and does not require special equipment or expertise. These attributes make the CIM more suited for clinical microbiology laboratories than other phenotypic methods.
Immunochromatographic assays
Many immunochromatographic (IC) assays have been recently developed and commercialized for rapid and precise detection of the primary carbapenemase enzymes from bacterial cultures. These tests use antibody-antigen binding-based technology to capture specific carbapenemase antigens. In 2011, Kitao et al52 introduced new IC test to detect MBL (IMP) producers in P. aeruginosa and Acinetobacter. This rapid assay detected all IMP-producers, consistent with PCR results. Moreover, the KPC-k-set and OXA-48 K-set assays, for example, are commercialized forms of IC tests; both rely on monoclonal antibodies that specific to capture of epitopes specific for KPC and OXA-48-like enzymes using colloidal gold nanoparticles inked to a nitrocellulose membrane in a lateral flow device.53,54 Glupczynski et al54 evaluated the KPC-k-set and OXA-48 K-set tests. These authors found that they exhibited good performance with high accuracy for the rapid detection of KPC and OXA-48 enzymes. In another study, Wareham et al53 evaluated the OXA-48 K-set test on 82 enterobacterial isolates that were resistant to carbapenems. These authors concluded that the OXA-48 K-set identified all OXA-48 producers with 100% sensitivity and specificity within 10 mins. Another interesting feature of the IC assay is its ability to detect some allelic variants of OXA-48 such as OXA-204, OXA-244, OXA-181 and OXA-232.54 Very recently, Riccobono et al55 Nodari et al56 and Wareham et al57 demonstrated that the KPC-k-set and OXA-48 K-set tests also detected KPC and OXA-48 producers directly from clinical samples (e.g blood culture and rectal swabs). Immunochromatographic assays could be useful as a rapid and inexpensive screening method for countries that have attained a high endemic level of OXA-48 (e.g., the Middle East) and KPC (e.g., North and South America) (Figure 1).
Nonphenotypic-based methods. MALDI-TOF MS
Matrix-assisted laser desorption/ionization-time-of-flight mass spectrometry (MALDI-TOF MS) is extensively used to identify many pathogens based on the molecular weight of different chemical compounds of bacterial and fungi cells. Recently, MALDI-TOF has been used by many clinical laboratories to detect antimicrobial resistance (e.g., β- lactamase activity among gram-negative bacteria).58-61 The identification of β-lactamase activity using this method is based on measuring the degradation products of specific β-lactams (in the case of carbapenemase hydrolysis) when the extracted proteins of bacteria are incubated (typically for 2–4 h) with carbapenems (meropenem or ertapenem).30 The first MALDI-TOF carbapenemase assay was introduced by Hrabak et al. in 2011.58 This method was validated on Enterobacteriaceae and P. aeruginosa strains that only produced carbapenemases (VIM, IMP, KPC, and NDM). Although false-positive and false-negative results were reported in P. aeruginosa, this method exhibited a sensitivity of 96.67% and a specificity of 97.87%.58 Another MALDI-TOF carbapenemase assay exhibited a low sensitivity (approximately 76%) due to some difficulties with the detection of OXA-48-like enzymes.46 These authors also reported that the addition of ammonium bicarbonate to the reaction mixture improved its sensitivity to 98%.46 In addition, many studies have reported the option of using MALDI-TOF for the direct detection of carbapenemase-producers from blood cultures.61,62 In 2014, Lau et al63 introduced a second MALDI-TOF technique based on the detection of the plasmid-associated protein conferring carbapenem resistance. This technique has only been used to detect the blaKPC carbapenemase-bearing pKpQIL plasmid in CRE isolates that were responsible for an outbreak at the National Institutes of Health (NIH) Clinical Center in 2011, in the United States.63 Although MALDI-TOF carbapenemase assays appear more attractive for routine work because MALDI-TOF apparatuses are available in many clinical laboratories, these methods remain too expensive for some clinical laboratories particularly those located in developing countries. Moreover, like many phenotypic carbapenemase detection methods, MALDI-TOF assays have difficulties in detecting specific carbapenemase (e.g., OXA-48). Remarkably, false-negative results have been reported and have been attributed to longer incubation times, which may lead to further decompositions of the degradation products to a very light mass. Such decompositions can render the products undetectable by a MALDI-TOF assay.21,58 Finally, there is no doubt that MALDI-TOF is a very useful microbial identification technology. However, its routine use for carbapenemase detection requires expert users to use MALDI-TOF settings other than those recommended for bacterial identification.30
Molecular detection of carbapenemase genes
Molecular techniques are still gold standard and more reliable for detecting carbapenemase-producing bacteria than phenotypic methods.34 These techniques include PCR-based methods, hybridization-based techniques and WGS.
PCR-based methods
Most of the molecular techniques for detecting carbapenem-resistance are PCR-based methods. They are used as a reference method to confirm or to address the problems in some results obtained from phenotypic detection methods. These approaches, either uniplex- or multiplex-PCR, are largely based on the amplification of a specific target region in chromosomal DNA. Polymerase chain reaction techniques can be performed on bacterial colonies with turnaround times of 4-6 h or directly on clinical specimens in the case of real time-PCR, which can yield a result within 1 h.34 A plethora of multiplex-PCR techniques have been developed for both laboratory-developed tests (LDTs) and commercial assays. Ellington et al64 (2007) introduced a multiplex-PCR technique for the identification and differentiation of the alleles encoding 5 different families of acquired MBL genes (IMP, GIM, VIM, SPM, and SIM). This method exhibited good performance in terms of detecting and ensuring allelic discrimination of all five families of MBL genes including the IMP and VIM variants. In 2011, another multiplex-PCR assay was developed by Poirel et al. to detect 11 carbapenemases genes using three different multiplex reactions. These genes belong to different classes and include the clinically most important carbapenemase genes (IMP, VIM, NDM, KPC, SPM and OXA-48) and less clinically important “minor” carbapenemase genes (AIM, DIM, GIM, SIM and BIC). Poirel et al. evaluated this method and demonstrated good performance against many control and clinical strains of carbapenemase-producing gram-negative bacilli (e.g., E. coli, K. pneumoniae, Enterobacter cloacae, P. aeruginosa, A. baumannii and Citrobacter freundii).65 In the last decade, multiplex real-time-PCR has become more attractive for detecting carbapenemase genes due to its ability to conduct simultaneous detection and differentiation of many carbapenemase types with a short turnaround time (≤60 min). The first multiplex real time-PCR method was developed by Mendes et al66 (2007) for detecting MBL-encoding genes (VIM, IMP, SPM-1, GIM-1 and SIM-1). The characterization of these carbapenemases was based on different amplicon sizes and melting peak temperatures (Tm). This method was excellent at detecting all MB-harboring clinical isolates. Another in-house, multiplex real time-PCR method was introduced to detect 6 different carbapenemases types (KPC, NDM, IMP, VIM, GES and OXA-48) that had been already identified in Enterobacteriaceae in a single reaction. This assay was tested against 30 well-known isolates carrying those six carbapenemase genes; the concordance was 100% with previous genotyping results.67 In 2014, van der Zee et al68 developed another real time-PCR-based test for detecting KPC, NDM, VIM, IMP, and OXA-48. This assay was evaluated by seven different laboratories and exhibited 100% sensitivity and specificity. This assay appears to be useful for detecting the majority of the dominant carbapenemase genes.68 Recently, several commercial multiplex PCR-based techniques have been developed and shown to have sensitivities between 97 and 100%.30,69-71 These assays include hyplex SuperBug ID and the Xpert Carba-R assay (for the detection of KPC, NDM, VIM, IMP and OXA-48)69,71 and Check Direct CPE (for KPC, VIM, NDM and OXA-48).70 These commercial assays are useful tools for carbapenemase detection from surveillance cultures, which can result in early implementation measures for infection control. It is clear that PCR-based methods are rapid, robust and reliable for the specific detection of the most dominant carbapenemase families. However, in addition to high costs and specific expertise and equipment required to perform these tests, most of the current PCR-based methods cannot detect new carbapenemase types or new variants of known types.
Hybridization-based techniques (microarrays)
A microarray format is a hybridization-based technique that allows for the detection of a large number of genes within a single reaction. Microarrays (also known DNA chips) are microscopic DNA spots with a unique DNA sequence (called probes). The DNA of bacteria is hybridized with complementary base pairs of probes in the slides and then scanned to see only probe-target hybridization. Microarrays have been commonly used for the identification and characterization of many bacterial species.72-74 This technology has also been used to detect and identify many carbapenemase genes. In 2010, a microarray assay was developed by Ulyashova et al. to identify seven different carbapenemase types (KPC, VIM, OXA, IMP, SPM, SIM, and GIM). This method involves several steps, including a simultaneous amplification of the β-lactamase genes using multiplex PCR followed by hybridization of oligonucleotide probes with labeled DNA. This method was evaluated on A. baumannii, P. aeruginosa, E. coli, and K. pneumonia strains that produce KPC-3, VIM-1, VIM-2, VIM-4, VIM7, IMP-2, IMP-1, SPM-1, OXA-40, OXA-23, OXA-51 and OXA-58. The researchers concluded that this microarray assay accurately identified all of the carbapenemase genes in the strains of interest.75 Recently, many commercial microarray-based assays have been developed including the Verigene gram-negative blood culture nucleic acid test (BC-GN), BioFire, and Check-Points. The Verigene (BC-GN), for example, was developed to detect the eight most common bacterial pathogens directly in blood cultures and six key β-lactamase types (IMP, KPC, NDM, OXA, VIM and CTX-M). The Verigene (BC-GN) assay was evaluated by some studies and found to exhibit excellent sensitivity and specificity.30,34,76,77 The advantage of a microarray assay over PCR-based techniques is in the number of available targets for investigation; PCR can target roughly 4-5 DNA sequences per assay, and microarrays can detect hundreds or thousands of targets.30 Despite this advantage, microarrays have some limitations such as being labor-intensive and associated with relatively high costs, which restrict their routine use in clinical laboratories.34
WGS
The sequencing platforms that were developed in the 1970s by Sanger, are based on the amplification of DNA by a DNA polymerase enzyme using normal deoxy-nucleotides triphosphates (dNTPs) and chain-terminating dideoxynucleotides triphosphates (ddNTPs) that are labeled with 4 different fluorophores.78 The resulting fragments can be analyzed by mass spectrometry or capillary electrophoresis.78,79 Sanger sequencing has been used in some clinical laboratories to investigate the sequencing of many antibiotic resistance genes. Nevertheless, conventional Sanger sequencing technology is labor intensive, time consuming, expensive, and inefficient for larger-scale projects such as WGS.78 Whole-genome sequencing is a next-generation sequencing (NGS) technology and is the most comprehensive molecular technology that provides all of the genetic information about an organism of interest in a cost-effective way. The ability of NGS to analyze DNA along the entire genome in a short time at relatively low costs renders NGS superior to the Sanger method. Whole-genome sequencing has already used for epidemiological investigations in some recent outbreaks, including Escherichia coli O104:H4,80,81 Vibrio cholera,82 Salmonella enteric,83 methicillin-resistant Staphylococcus aureus (MRSA),84 and carbapenem-resistant K. pneumoniae.85 Additionally, WGS has become an important tool for the development of new antibacterial agents by enabling the rapid identification of all known resistance mechanisms including carbapenemases. Furthermore, WGS can identify other mechanisms of resistance such as porin mutations and types of plasmid-carrying resistance genes.86 Whole-genome sequencing data can also provide a rout to future inquiry when novel resistance mechanisms or virulence determinants of interest are identified.30 However, WGS is still comparatively expensive and necessitates automated pipelines for data analysis.
Conclusion
Although enormous advancements in medical technology and services have occurred within the last two decades, antibiotic resistance has dramatically increased worldwide. Therefore, some scientists have warned that if antibiotic resistance continues as it has the world could return to the pre-antibiotic era and multidrug-resistant (MDR) infections could result in roughly 10 million deaths by 2050.87 Consequently, the development of new strategies to face this imminent danger has become an urgent priority for the World Health Organization (WHO) and many governments.88 These strategies include stricter regulations on antibiotic use to minimize the emergence of antibiotic resistance and the dedication of sufficient research funds for novel antibacterial drug development. Undoubtedly, rapid detection and characterization of antibiotic-resistant bacteria using reliable techniques play an essential role in both the treatment of MDR infections and controlling the further spread of those isolates. Nowadays, the detection and characterization of CROs represent a true challenge for many clinical laboratories around the world. In this review, the various phenotypic and non-phenotypic methods for the detection of CROs were discussed. No consensus has yet been reached with regards to a single optimal method. When selecting an ideal detection method for diagnostic use or screening purposes, it is important to consider many traits such as rapidity, cost, accessibility, ease of use and degree of accuracy (Table 2). Moreover, the sorts of resistance determinants, the prevalence of resistant bacteria, the geographical region, and the patient population are also important to consider when selecting and implementing a method. Phenotypic methods such as culture-based methods and screening media are simple and affordable, and they are more attractive for routine use in clinical laboratory settings. However, their sensitivity and specificity are still inconclusive, and they require long incubation times. Although some biochemical tests such as the Carba NP test exhibit low sensitivity to some carbapenemases such as OXA-48, these tests are sometimes preferable given their rapidity and simplicity. Immunochromatographic assays have recently garnered more attention because they can rapidly detect carbapenemase-producers directly from clinical specimens with a high level of accuracy. Due to the differences in carbapenem-resistant activity between carbapenemases that can be detected by different methods, the multiple use of two or more phenotypic detection methods can accordingly improve the detection of CROs compared with using a single technique. Molecular methods have become the method of choice because the majority can be performed rapidly with a high level of sensitivity and specificity. Molecular methods also possess other advantages over phenotypic methods such as enabling direct detection of CPOs from clinical specimens and permitting definitive identification of the exact mechanism of carbapenem resistance.21 These advantages are particularly useful not only for surveillance and epidemiological purposes but also for outbreak investigations.30 Nevertheless, some of these molecular techniques are unable to detect novel unidentified resistant genes; they are considered to be reference methods for confirmation and further characterization of known carbapenemases. They can only be used in a reference laboratory. Whole-genome sequencing yields unambiguous data that enable a complete analysis of the whole genome. We expaect that WGS may ultimately become a highly powerful tool for both outbreak investigations and molecular characterization of antibiotic resistance genes in routine clinical settings. However, WGS is still relatively expensive and requires an automated data interpretation system and a publicly available database. The routine implementation of WGS in clinical laboratories may not occur for several years in the future, particularly in developing countries.
Simplified comparison of performance characteristics of various phenotypic and molecular methods for the detection of carbapenemase-producing gram-negative bacilli.
Footnotes
Disclosure. Authors have no conflict of interests, and the work was not supported or funded by any drug company.
- Received May 12, 2018.
- Accepted July 25, 2018.
- Copyright: © Saudi Medical Journal
This is an open-access article distributed under the terms of the Creative Commons Attribution-Noncommercial-Share Alike 3.0 Unported, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.