Abstract
Objectives: To investigate the frequency of the growth hormone receptor (GHR)-d3 polymorphism in a random sample of Saudi Arabian population from Jazan province, and test the effects of the polymorphism on some anthropometric factors.
Methods: This cross-sectional population-based study was conducted during the period from January to April 2017 at the College of Applied Medical Sciences, Jazan University, Southwestern Saudi Arabia. A total of 230 healthy adult male and female volunteers were randomly recruited. Genomic DNA was extracted from the whole blood, and the GHR exon 3 locus was genotyped using multiplex polymerase chain reaction.
Results: The distributions of the GHR genotypes were as follows: fl/fl (39.1%), fl/d3 (44.8%), and d3/d3 (16.1%). No statistically significant differences were found between fl/fl, fl/d3, or d3/d3 GHR genotypes in terms of weight (p=0.90), height (p=0.12), or body mass index (BMI) (p=0.83) values.
Conclusion: No correlations were found between the GHR-d3 polymorphism and weight, height, or BMI.
Human growth hormone (hGH; somatotropin) is a peptide hormone synthesized in the adenohypophysis to promote growth-related cellular responses.1 Human growth hormone signaling is transduced by growth hormone receptor (GHR), which belongs to type I cytokine receptor superfamily.2 Full-length GHR is encoded by 9 exons located on the short arm of chromosome 5, region 5p13-p12.3 Exons 3-9 are translated into full-length GHR, which is a single pass transmembrane protein composed of 246 amino acid residues for the extracellular domain and short transmembrane domain and 350 amino acids residues for the intracellular domain.3 Deletion of exon 3 from the GHR gene (GHR-d3) is a common human-specific polymorphism that occurred several million years ago as a result of homologous recombination.4 This results in 3 possible GHR gene allelic variants: fl/fl, in which both alleles have the full-length copy of the GHR gene; fl/d3, in which one allele carries the full-length copy and the other carries the exon 3 deletion mutation; and d3/d3, in which both alleles carry the exon 3 deletion mutation. Deletion of exon 3 results in elimination of 23 amino acids from the GHR extracellular domain, which influences GHR membrane trafficking and stability, but does not alter the GH/GHR interaction.5,6 The clinical relevance of the GHR-d3 polymorphism remains controversial. In a meta-analysis study, a modest association between the GHR-d3 gene and increased responsiveness to exogenous recombinant human GH (rhGH) therapy in short children with GH deficiency, small for gestation age (SGA), or Turner syndrome has been reported,7 although some studies have reported that there is no association.8,9 The GHR-d3 polymorphism has also been shown to influence the body composition under some conditions. For example, this polymorphism has been linked to increased insulin secretion and higher triglyceride levels in normal individuals at puberty.10 Associations with increased body mass index (BMI) and insulin resistance have been reported in acromegalic patients.11,12 In contrast, the GHR-d3 polymorphism was found to be associated with lower BMI in obese children13 and in girls with Turner syndrome.14 Some studies have also reported a potential impact of the GHR-d3 genotype on height.15,16 Kang et al15 reported an association between the GHR-d3 polymorphism and mandibular bone height. A study in a population of Greek children17 reported a significant height gain and growth velocity in GHR-d3 carriers who received rhGH therapy for one year.
The aim of this work was to investigate the prevalence of different GHR genotypes among the Saudi Arabian population and evaluate the influence of such polymorphisms on weight, final height, and BMI.
Methods
This was a cross-sectional study involving 230 healthy Saudi Arabian volunteers (144 men and 86 women) from Jazan province. The individuals were recruited from the College of Applied Medical Sciences, Jazan University, southwestern Saudi Arabia, between January and April 2017. The inclusion criteria were as follows: Saudi Arabian citizens from Jazan Province and age between 20 and 45 years. The exclusion criteria were as follows: history of obesity, growth-related disorders, or chronic diseases.
All participants were asked to read and sign an informed consent form. The study was approved by the research ethical committee of King Fahd Central Hospital in Jazan. Before blood sample collection, demographic data, including gender, age, weight, and height, were collected. The BMI was calculated from weight in kilograms (kg) and height in meters (m) according to the formula: BMI = weigh (kg) / height (m)2.
Blood collection and DNA extraction
Whole-blood samples were collected from each subject in 5-mL ethylenediaminetetraacetic acid vacutainer tubes using an aseptic phlebotomy technique. For genomic DNA extraction, 200 µL of the collected blood samples was applied to an Illustra Blood Genomic Prep Mini Spin Kit (GE Healthcare Life Sciences Ltd., UK), according to the manufacturer’s instructions.
Genotyping
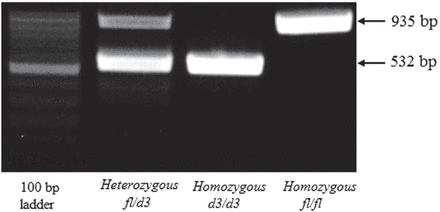
The full-length GHR allele (fl) and exon 3-deleted GHR allele (d3) were detected by multiplex polymerase chain reaction (PCR). Approximately 10 ng of the extracted DNA was used as a template for PCR with previously reported primers, as follows: (G1) forward primer, 5¢-TGTGCTGGTCTGTTGGTCTG-3¢, (G2) reverse primer, 5¢-AGTCGTTCCTGGGACAGAGA-3¢, and (G3) reverse primer, 5¢-CCTGGATTAACACTTTGCAGACTC-3¢ (GenBank accession no. AF155912). Polymerase chain reaction was carried out with a final volume of 25 µL. The reaction mixture contained 1 µL DNA, 1 µL of each primer, 8.5 µL sterile nuclease-free deionized distilled water, and 12.5 µL of 2× Top Taq Master Mix (Qiagen, Germany). The thermal protocol for PCR was as follows: initial denaturation for 4 min at 94°C followed by 35 cycles of 30 seconds of denaturation at 94°C, 30 seconds of annealing at 57°C, and 45 seconds of elongation at 72°C. After the cycles, a final extension step was carried out at 72°C for 10 min. The PCR products were then run on 1% agarose gel and stained with ethidium bromide for DNA visualization and imaging. The fl allele was represented by a 935-bp fragment, and the d3 allele was represented by a 532-bp fragment on the agarose gel. The homozygous full-length genotype (fl/fl) was indicated by the presence of a single band at 935 bp, whereas the homozygous exon 3-deleted genotype (d3/d3) was indicate by the presence of a single band at 532 bp. Heterozygous carriers of the full-length and exon 3-deleted genotype (fl/d3) were represented by the presence of 2 bands at 935 and 532 bp on the agarose gel (Figure 1).
Polymerase chain reaction products on 1% agarose gel showing a representative examples of all 3 possible growth hormone receptor (GHR), fl/d3 allelic variants. The homozygous full-length genotype (fl/fl) indicated by the presence of a single band at 935 bp, while the homozygous exon 3 deleted genotype (d3/d3) indicate by the presence of a single band at 532 bp. The heterozygous carriers of the full-length and exon 3 deleted genotype (fl/d3) represented by the presence of 2 bands at 935 and 532 bp.
Statistical analysis
Hardy-Weinberg equilibrium (HWE) between the observed and expected GHR genotype frequencies was calculated using Chi-squared (χ2) tests. Anthropometric data were expressed as percentages or means ± standard deviations (SDs). Height, weight, and BMI were first transformed into SD scores (SDSs) according to age- and gender-matched national standards. Comparisons between means of different GHR genotypes were carried out using one-way analysis of variance (ANOVA). Differences were considered statistically significant if the P value was less than 0.05. All statistical analyses were conducted using GraphPad Prism software (San Diego, CA, USA).
Results
The distribution of all GHR genotypes in the present study were in Hardy-Weinberg equilibrium (Table 1). Analysis of the distribution of the GHR genotypes in the total study population showed that 39.1% of subjects carried the fl/fl genotype, 44.8% carried the fl/d3 genotype, and 16.1% carried the d3/d3 genotype (Table 2). Single allele frequencies were 61.5% for fl and 38.5% for d3 allele. Analysis of variance showed that there were no significant differences in weight, height, and BMI SDSs across different GHR genotypes (p=0.90, p=0.12, and p=0.83).
Growth hormone receptor-d3 allele frequencies and Hardy-Weinberg equilibrium.
Baseline characteristics and the GHR genotypes of the study population (n=230)
Discussion
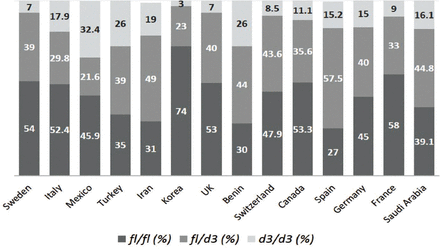
Growth hormone receptor-d3 is a common polymorphism in humans and has been found to alter responses to rhGH therapy in GH-deficient patients and to affect newborn infant size, glucose metabolism, and BMI.18 The prevalence of the GHR-d3 polymorphism in the Saudi Arabian population and its association with basic anthropometric parameters, such as height and BMI, have not been studied. In this study, the prevalence of this polymorphism in men and women from Saudi Arabia (Jazan province) and the correlations with anthropometric measurements were investigated as a preliminary step for further exploratory studies on the clinical relevance of such polymorphisms in the Saudi population. The findings showed that 39.1% of subjects carried the GHR-fl/fl genotype, 44.8% carried the fl/d3 genotype, and only 16.1% carried the homozygous mutated genotype d3/d3. The closest country to our overall GHR allelic distributions is Iran, with fl/fl of 31%, fl/d3 of 49%, and d3/d3 of 19%.19 As reported in some countries, such as Spain, Benin, and Turkey, we found higher prevalence rates of the GHR fl/d3 genotype than the other genotypes.11,20,21 However, in some European and western countries, such as the UK, France, Germany, Sweden, Switzerland, and Canada, higher prevalence rates of the GHR fl/fl genotype have been observed.16,22-26 The frequency of the homozygous GHR d3/d3 genotype was 16.1%, which was similar to the average frequencies found in Italy (17.9%), France (19%), and Germany (15.2%).22,23,27 Moreover, the highest frequency of the GHR d3/d3 genotype was reported in Mexicans (32.4%), and the lowest was reported in Koreans (3%)28,29 (Figure 2).
The frequency of the growth hormone receptor (GHR) exon 3 genotypes in different populations in comparison with the present study in Saudi Arabia. fl/fl - homozygous full-length genotype, d3/d3 - homozygous exon 3 deleted genotype, fl/d3 - heterozygous carriers of the full-length and exon 3 deleted genotype
Consistent with several reports, no correlations were observed between the GHR-d3 polymorphism and weight, height, or BMI in this study based on the healthy adult population.16,21,30,31 Nevertheless, some studies have revealed an association between the GHR-d3 polymorphism and reduced BMI under some conditions. For example, a Chinese study conducted on obese children reported significant correlations of GHR d3/d3 with low BMI, low insulin resistance, and low total cholesterol.13 Similarly, Binder et al14 reported a low BMI in patients with Turner’s syndrome with homozygous GHR d3/d3 alleles.
The current study has some limitations, including the relatively small sample size compared with the total Saudi Arabian population. Additionally, the study was conducted locally in one region of the country, and samples were collected only from healthy subjects; the GHR-d3 polymorphism has not been linked to some clinical outcomes, such as short stature.
In conclusion, the findings of this study represent the first reported distribution of GHR gene variants among Saudi Arabians. The distributions were 39.1% for the fl/fl genotype, 44.6% for the fl/d3 genotype, and 16.1% for the d3/d3 genotype. Consistent with several reports, no correlations were found between the GHR-d3 polymorphism and weight, height, or BMI in this study. However, further studies are required to demonstrate the contribution of the GHR-d3 polymorphism to birth size, BMI, and final height in Saudi Arabians and other Gulf countries.
Withdrawal policy
By submission, the author grants the journal right of first publication. Therefore, the journal discourages unethical withdrawal of manuscripts from the publication process after peer review. The corresponding author should send a formal request signed by all co-authors stating the reason for withdrawing the manuscript. Withdrawal of a manuscript is only considered valid when the editor accepts, or approves the reason to withdraw the manuscript from publication. Subsequently, the author must receive a confirmation from the editorial office. Only at that stage, are the authors free to submit the manuscript elsewhere.
No response from the authors to all journal communication after review and acceptance is also considered unethical withdrawal. Withdrawn manuscripts noted to have already been submitted or published in another journal will be subjected to sanctions in accordance with the journal policy. The journal will take disciplinary measures for unacceptable withdrawal of manuscripts. An embargo of 5 years will be enforced for the author and their co-authors, and their institute will be notified of this action.
Acknowledgment
The author would like to take this opportunity and thank all volunteers who were exceptionally cooperative in the completion of this study. The author would also like to thank the deanship of scientific research at Jazan University, Jazan, Kingdom of Saudi Arabia for moral and financial support.
Footnotes
Disclosure. Authors have no conflict of interests, and the work was not supported or funded by any drug company.
- Received May 15, 2017.
- Accepted July 25, 2017.
- Copyright: © Saudi Medical Journal
This is an open-access article distributed under the terms of the Creative Commons Attribution-Noncommercial-Share Alike 3.0 Unported, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.