Abstract
Objectives: To elucidate the contribution of x-ray repair cross-complementing (XRCC) protein 1 399Gln, XRCC3 241M, and XRCC3-5’-UTR polymorphisms to the susceptibility of breast cancer (BC) in a Jordanian population.
Methods: Forty-six formalin fixed paraffin embedded tissue samples from BC diagnosed female patients, and 31 samples from the control group were subjected to DNA sequencing. Samples were collected between September 2013 and December 2014.
Results: The XRCC1 Arg399Gln genotype did not exhibit any significant correlation with the susceptibility of BC (odds ratio [OR]=1.45, 95% confidence interval [CI]: 0.60-3.51) (p=0.47). Likewise, XRCC3 M241T genotype did not show significant correlation with BC (OR=2.02, 95% CI: 0.50-8.21) (p=0.40). However, distribution of XRCC3-5’UTR (rs1799794 A/G) genotype showed a significant difference between the patient and control group (OR=0.73, 95% CI: 0.06-8.46) (p=0.02).
Conclusion: The XRCC3-5’UTR (rs1799794) G allele frequency was higher in cancer patients while XRCC1 (rs25487) and XRCC3 (rs861539) did not show any significant correlation with susceptibility of BC in the selected Jordanian population. Contribution of other environmental factors should be studied in future works, as well as the response of cancer therapy.
Breast cancer (BC) incidence in Jordan has been estimated at 1,237 cases in 2012, with a prevalence of 4,260 cases over 5 years, and mortality rate up to 426 cases.1 Genetic predisposition contributes to less than 10% of BC cases, which raises a demand for further research into new genetic markers of BC risks.2 Fewer than 5% of BC cases have been found to be mutated at breast cancer 1 (BRCA1) early onset and BRCA2 genes, and approximately 40% of familial BC families have been identified for genetic predisposition.3 Unfortunately, mammalian cells are habitually exposed to genotoxic agents, such as ionizing radiation that can lead to DNA damage. Many double strand break,4 and single strand break (SSB) repairing proteins have been identified including DNA repair protein homolog, or RAD tecombinase, or x-ray repair cross-complementing (XRCC)s family proteins.5 Deficiency in repairing system might contribute to cancer development due to the loss of genetic integrity and genome instability.6 Mutation in DNA repair proteins is very rare.7 Therefore, many studies have been conducted to evaluate the role of allelic polymorphisms in DNA repair genes involved in cancers development.8,9 Genetic polymorphisms in DNA repair genes XRCC1, and XRCC3 have been screened to find an association with the risk of BC.10-12 Studies have demonstrated an association between XRCC1 and XRCC3 polymorphisms, and certain cancers subsuming colorectal cancer,13 lung cancer,14 pancreatic cancer,15 head and neck cancer,16 gastric cancer,17 esophageal cancer,18 melanoma skin cancer,19 oral squamous cell carcinomas,20 lung cancer risk,21 bladder cancer,22 and BC.23 Furthermore, a meta-analysis study supported the contribution of XRCC1 Arg399Gln polymorphism in susceptibility of BC in the American population.24 On the other hand, no relationship has been found between XRCC1 and XRCC3 polymorphisms and the risk of BC,25 lung cancer,26 bladder cancer,27 prostate cancer,28 lung cancer risk,29 cutaneous malignant melanoma,30 furthermore, it may decrease the risk for myeloblastic leukemia31 and non-melanoma skin cancer.32 Alcoholism, abortion, and non-breast feeding have been associated with increased risk of BC with contribution of XRCC1 399Gln and XRCC3 T241M polymorphisms.11 Moreover, family history,12 age group,33 polycyclic aromatic hydrocarbon-DNA adducts, fruit and vegetable and antioxidant intake, and non-smokers have been suggested to be associated with the risk of BC in interaction with XRCC1 or XRCC3 polymorphisms.34 The aim of the current study was to elucidate the contribution of XRCC1 399Gln, XRCC3 241M and XRCC3-5’-UTR polymorphisms in the susceptibility of BC in the Jordanian population. This study is intended to establish a reference point for future single nucleotide polymorphism (SNP) studies in the Jordanian population, which may contribute to the development of a national cancer database.
Methods
Forty-six formalin fixed paraffin embedded tissue blocks (FFPE) were collected randomly from sporadic BC female patients between September 2013 and December 2014 at the King Abdullah University Hospital, Irbid, Jordan. All samples were chosen to be triple negative. A control group of 31 blood samples were collected from age matched normal Jordanian females without a diagnosis of BC. This study was conducted according to the principles of Helsinki Declaration.
DNA extraction
Ten-µm thickness of 4 sections of FFPE samples were deparaffinized by 2 steps of Xylene (Sigma-Aldrich Corp., St. Louis, Missouri, USA) for 5 minutes each, followed by centrifugation for 5 minutes at 1,6000 g (Microcentrifuge 5415D, Eppendorf, Germany). Xylene was cleared by 2 steps of absolute ethanol; by incubation for 5 minutes each step followed by centrifugation at 1,6000 g. ethanol (Sigma-Aldrich Corp., St. Louis, Missouri, USA) was evaporated by air drying in the chemical cabinet for 20-30 minutes. Genomic DNA from tumor tissues and corresponding control samples were prepared according to the manufacturer’s instructions Qiagen DNeasy kit (Qiagen GmbH, Hilden, Germany).
Polymerase chain reaction (PCR) amplification and DNA sequencing
The PCR amplifications targeting the XRCC1-exon-10, XRCC3-exon-7 and XRCC3-5’-UTR regions were performed using specific primers based on the XRCC1 and XRCC3 sequences obtained from the National Center for Biotechnology Information (NCBI) (Table 1). The PCR amplification was performed in 30 µl reaction volume that contained (75 mM Tris-HCl, 1.5 mM MgCls, 50 mM KCl, 20 mM (NH4) 2SO4, 0.2 mM of each primer and 1 U of Taq DNA polymerase). Polymerase chain reactions were conducted under the following cycling conditions: an initial 7 minutes of denaturation at 95°C followed by 45 cycles for 45 seconds each at 94°C, 59°C, 72°C for 1 minute, and a single final extension step for 10 minutes at 72°C. Direct DNA sequencing was performed using Big Dye Terminator version 3.1 kit (Applied Biosystems, Waltham, MA, USA). Samples were run on an ABI Prism Genetic Analyzer system 3130xl (Applied Biosystems, Waltham, MA, USA).
Primer pairs of selected sequences of the target gene areas with their corresponding melting points.
Statistical analysis
Fisher’s exact test analysis was used for the calculation of p-value, odds ratio (OR) and 95% confidence interval (CI) and Hardy-Weinberg Equilibrium (HWE) evaluation. GraphPad Prism-6 software was used for statistical analysis. P<0.05 was considered significant.
Results
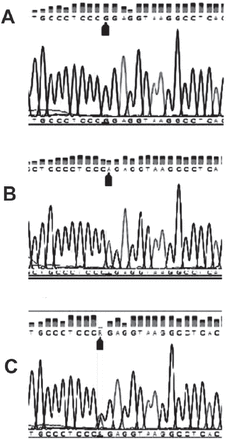
The mean diagnostic age of cancer patients was 54 years. Genotypes of the selected genes were confirmed by direct sequencing as shown in Figure 1. Allele frequencies in controls were consistent with Hardy-Weinberg Equilibrium (p=0.24 for Arg399Gln, p=0.33 for M241T, and p=0.97 for XRCC3-5’UTR). Genotypic distributions of the 3 positions of the selected genes are shown in Tables 2-4. Frequencies of XRCC3-5’UTR (rs1799794 A/G) genotype showed a significant difference between the patients and control group (OR=0.73, 95% CI: 0.06-8.46) (p=0.02). On the other hand, XRCC1 Arg399Gln genotype did not exhibit any significant association with the susceptibility of BC (OR=1.45, 95% CI: 0.60-3.51) (p=0.47). Likewise, screening of XRCC3 M241T genotype in cancer samples did not show a significant difference in comparison with the control group, which supports the weak association with BC risk (OR=2.02, 95% CI: 0.50-8.21) (p=0.40).
A DNA sequencing of x-ray repair cross-complementing family proteins (XRCC)1-Arg399Gln (rs25487) single nucleotide polymorphism showing: A) homozygous CGG (Gln); B) homozygous CAG (Arg); and C) heterozygous CGG/CAG (Gln/Arg).
Genotype distribution of x-ray repair cross-complementing protein (XRCC)1 Gln399Arg (rs25487).*
Genotype distribution of x-ray repair cross-complementing protein (XRCC)3 Met241Thr (rs861539).*
Genotype distribution of x-ray repair cross-complementing protein (XRCC)3-5’UTR-A/G (rs1799794).*
Discussion
Genetic mutations in DNA repair genes are very rare in cancers; therefore, many studies have been conducted to evaluate the role of genotypic polymorphisms of many DSB and SSB repairing proteins in the development of cancer.5 Controversially, some studies have found significant correlation between certain polymorphisms of XRCC1 and XRCC3 genes and the susceptibility of various cancers including breast cancer.16,20,24,35 On the other hand, other studies have not found any significant relationship between these polymorphisms and cancer development.36,37 Contributions of other factors have been suggested in the involvement of cancer risk with XRCCs genes polymorphisms, such as age, family history, smoking, diet and alcoholism.11
The present study did not find any significant relationship between XRCC1 Arg399Gln and XRCC3 M246T genotypes and breast cancer risk, which is consistent with the findings of other studies.25 The Arg399Gln has been found to be involved in increasing BC risk among Asians (OR - 1.26, 95% CI: 0.96-1.64), and Africans (OR - 1.80, 95% CI: 0.97-3.32). In addition, it is associated with a slight increase in BC risk in Caucasians (OR - 1.08, 95% CI: 0.95-1.22).36 The current results showed a significant relationship between XRCC3-5’UTR A/G (rs1799794) genotype and BC risk. The XRCC3-5’UTR-G allele frequency was more common in cancer patients in comparison to the control group. The later polymorphism is consistent with a meta-analysis study.38 Functional studies are required to understand the role of XRCC3-5’UTR A/G (rs1799794) polymorphism in gene expression and cell proliferation.
This study does not exclude the role of XRCC1 (rs25487) and XRCC3 (rs861539) in the development of BC due to many reasons. Firstly, sample size needs to be bigger in the Jordanian population, which is one of the limitations of our study. Secondly, more clinicopathological data might have significant contribution with these polymorphisms. Finally, ethnic origin, smoking, alcoholism, abortion, non-breast feeding, and diet can be contributing factors.11 Moreover, XRCC1 (rs25487) and XRCC3 (rs861539) polymorphisms have been suggested to be involved in radiotherapy response and survival of BC patients.39 Furthermore, combination of variants of XRCC1 and XRCC3 genes has been suggested to be associated with susceptibility to BC with limited sample size.25 Moreover, a predictive value of XRCC1 (399Gln) and XRCC3 (241Met) polymorphisms in the survival of metastatic BC after radiotherapy and adjuvant chemotherapy treatment can be another benefit of such findings.39 The differences observed in the current and other studies may be due to limited sample size, type of samples (FFPE) and different genetic background.
In conclusion, XRCC3-5’UTR A/G (rs1799794) genotype showed a significant correlation with BC risk. Moreover, G allele frequency was higher in cancer patients, whereas XRCC1 (rs25487) and XRCC3 (rs861539) did not show any significant correlation with susceptibility of BC in the selected Jordanian population. Further studies are required to rule out or confirm such findings. Contribution of other environmental factors should be studied in future works, as well as the response of cancer therapy. Collection of blood samples from larger population would be more efficient for association with BC risk.
Acknowledgment
The author gratefully acknowledges Dr. Emad Malkawi, Dr. Alaa Al-Jabali, and Dr. Kalid Batayneh for their support and assistance in completing this study.
Footnotes
Disclosure. Author has no conflict of interests, and the work was not supported or funded by any drug company.
- Received June 11, 2015.
- Accepted September 4, 2015.
- Copyright: © Saudi Medical Journal
This is an open-access article distributed under the terms of the Creative Commons Attribution-Noncommercial-Share Alike 3.0 Unported, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.