Abstract
Objectives: To study serotype distribution and antimicrobial resistance to beta-lactams and macrolides in pneumococci causing respiratory diseases after the introduction of the 13-valent pneumococcal conjugate vaccine in Saudi Arabia.
Methods: This is a hospital-based and a cross-sectional prospective surveillance study conducted at King Fahad Hospital of the University, AlKhobar, Kingdom of Saudi Arabia, in which respiratory pneumococcal isolates collected between 2012 and 2014 were serotyped by multiplex sequential polymerase chain reaction (PCR) and Pneumotest-Latex. Resistance genes to beta-lactams and macrolides were detected by multiplex PCR.
Results: The most common serotypes encountered were 11A, 19A, 17F, 23F, 3, and 19F, representing 64% of the typeable strains. Interestingly, 24% of the 94 isolates were not typeable and 18% were negative for the housekeeping gene cpsA. Among the 53 typeable pneumococci isolates, 36 (67.9%) carried genes encoding resistance to both penicillin and macrolides, 9 (17%) were penicillin-monoresistant, 3 (5.6%) were macrolide-monoresistant, and 5 (9.4%) were designated non-resistant. The high rate of resistance genes did not significantly differ according to serotype (p=0.76). Similarly, non-typeable pneumococci (cpsA+ and cpsA-) had high rates of resistance to both penicillin (62.5%) and macrolides (47%).
Conclusion: These data highlight the emergence of a previously rare capsular type, 11A (mean patient age, 29 years; p=0.001). Moreover, the high percentage of non-typeable isolates shows the emergence of possible atypical pneumococcal serotypes not covered by available vaccines.
Streptococcus pneumoniae (S. pneumoniae) is known worldwide as a causative agent of community-acquired pneumonia, especially in children and the elderly. The capsule polysaccharide (CPS) that encapsulates the bacterial cell plays a key role in pneumococcal virulence. At present, more than 90 different polysaccharides have been identified and the sequences of all capsular loci have been determined.1 Pneumococcal invasive diseases are mainly caused by 15 serotypes (14, 6, 1, 19, 3, 4, 5, 9, 18, 23, 12, 7, 2, 25 and 8).2 Several pneumococcal vaccines have been used globally, including pneumococcal conjugate vaccine (PCV)7, PCV10, PCV13, and pneumococcal polysaccharide vaccine (PPV)23, which represent increasing coverage of the invasive serotypes. The non-conjugate vaccine PPV23, however, varies in the degree of immune response it elicits.3 A new conjugate vaccine, PCV15, is currently undergoing clinical evaluation.4 In Saudi Arabia, a new 13-valent vaccine (PCV13) was introduced in 2010; it covers the 13 types of pneumococci that most commonly cause pneumococcal infections, accounting for 91% of cases as reported by the Ministry of Health The incidence of invasive pneumococcal disease5 (IPD) in children <5 years, between 2007 and 2009, was estimated to be 2.5-21.6 per 100,000.6 Due to the high risk of IPD among children <5 years and seniors >50 years, the most recent Saudi guidelines recommend that all children receive 4 doses of PCV13, to be completed before the first year of age, and that seniors be vaccinated with PCV13, followed by PPSV23 one year later.7
Along with the virulent nature of the pathogen, the high genomic plasticity of S. pneumoniae makes its development of resistance to various classes of antimicrobials a huge problem, leading to high morbidity and mortality worldwide.8 Beta-lactams and macrolide antibiotics are widely prescribed to treat pneumococcal diseases. However, pneumoccocal resistance to these drugs has emerged as a worldwide issue since the first resistance to penicillin was documented in 1965. Since then, pneumococci have quickly acquired resistance genes and became increasingly resistant to antibiotics, causing treatment failures and consequently serious public health concerns. In Saudi Arabia, multidrug resistant pneumococci is becoming increasingly prevalent.9 Several different mechanisms underlie antimicrobial resistance in pneumococcus: mutation, transformation or upregulation of efflux pumps, and target modification.10-13 The mode of action of ß-lactams involves the transpeptidases, multiple enzymes active in cell wall biosynthesis; these enzymes are penicillin binding proteins (PBPs). Mutation of three PBP genes (pbp1a, pbp2b and pbp2x) leads to penicillin-resistance at a molecular level. The effects of some of these ß-lactam-resistant mutations can be overcome clinically by doubling the dose of the drug in pneumococcal respiratory diseases; however, this strategy is not effective in invasive infections.14 Macrolides resistance were encoded by 2 genes, ermB (responsible for 23S rRNA methylation) and mefA (macrolide efflux), and tends to be dose-independent.15 In resistance mediated by ermB, the bacterial ribosome to which the antimicrobial drug binds is modified by a new enzyme, which is coded by newly transferred DNA.16
In Saudi Arabia, multi-drug resistant pneumococci have been reported with resistance to multiple commonly used antimicrobial agents.10,17 A recent study has shown that more than 75% of invasive pneumococcal disease burden in Saudi Arabia can be attributed to the serotypes 23F, 6B, 19F, 18C, 4, 14, and 19A. However, little is known about alterations in the local epidemiology of respiratory pneumococcal diseases in the post-vaccination era.18 Surveillance of pneumococcal serotypes worldwide is important to understand the dynamics of pathogen transmission. In addition, identifying changes in serotype distribution among local pneumococcal strains provides evidence-based recommendations for the optimal implementation of future conjugate vaccines.
The aim of this prospective work was to study S. pneumoniae serotypes isolated from the Eastern Province of Saudi Arabia after routine use of the 13-valent pneumococcal conjugate vaccine (PCV13). This study also investigated trends in antimicrobial resistance to ß-lactams and macrolides, as well as the relationship between serotypes and antimicrobial resistance patterns.
Methods
This is a hospital-based and a cross-sectional prospective surveillance study conducted at King Fahad Hospital of the University, AlKhobar, Kingdom of Saudi Arabia. All S. pneumoniae isolates were collected between January 2012 and December 2014 from upper and lower respiratory tract specimens if they were thought to represent disease rather than colonization status. Ethical approval was obtained prior to the study and only good-quality lower respiratory tract specimens were included in the analysis (purulent portion with more than 25 leucocytes/low power field (lpf) and less than 10 epithelial cells per lpf). Only sputa suggestive of infection, with heavy dominating or pure pneumococcal growth, were included.
Bacteriological methods. Identification, storage and retrieval of pneumococcal strains
Colonial morphology on Columbia Blood Agar (CBA) and sensitivity to a 6-mm optochin disk (5 µg) were used to identify bacterial species after 24 hours of incubation. Disks were placed at the junction of the primary inoculum and second streak and incubated in 5% CO2 for 24 hours. Zone sizes were measured and interpreted in accordance with Clinical & Laboratory Standards Institute (<14 mm is resistant; ≥14 mm is susceptible). The identities of strains were then confirmed by Vitek II (bioMérieux, Paris, France) and both API-20 Strep and API-32 rapid Strep (bioMérieux, Paris, France), following the manufacturer’s recommendations, before being stored in brain heart infusion (BHI) broth with 12% glycerol at -80°C.
Quality control
As recommended by the CLSI, S. pneumoniae ATCC 49619 was used as the quality control (QC) strain for all tests. The supermaster stock was stored at -80°C in cryovials, while 2 master stocks were stored in 2 locations in a plastic storage box, sealed and clearly labeled in a -80ºC freezer. For retrieval, strains were removed from the freezer and cryovials were allowed to thaw slightly at room temperature. Using aseptic techniques, one loopful of culture was removed from the vial and placed on CBA using a sterile, disposable culture loop, and the plate was labeled with a pre-printed QC strain sticker. With another sterile loop, the plate was streaked to create an initial inoculum. Plates were then incubated at 37°C in 5% CO2. The cryovial with unused culture was returned to the -80ºC freezer as soon as subculturing had been completed. After 18 hours of incubation, the culture plates were examined to check for purity and that the morphological characteristics were consistent with those of the desired organism. Any mixed growth was re-subbed; if continual mixed growth occurred, the supermaster stock was used as a new starting point. If no growth occurred despite these procedures, the organism was considered nonviable. Whenever susceptibility testing was performed, the QC strain was included and the results were accepted only if they fit into the expected range. If the QC failed, investigations and corrective action were performed as per the CLSI Guidance.
Phenotypic serotyping
Serotyping of isolates was carried out based on the Pneumotest-Latex reaction, covering all known serotypes with a complete set of specific antisera in the Danish chessboard typing system (Statens Serum Institut, Copenhagen, Denmark), as recommended by the manufacturer and previously described.19 Briefly, latex agglutination was performed by mixing an aliquot of 10 µL of a pneumococcal broth culture (BHI broth, SPML, Saudi Arabia) with 10 µL of latex reagent on a microscope slide, and the slide was manually rocked for 5-8 seconds (s). A positive reaction with agglutination was read with the naked eye within 5-10 s if the latex reagent contained antiserum homologous to the capsule of the pneumococcal culture. Agglutinations observed after more than 10 s were considered nonspecific and thus ignored. Reactions with 14 latex reagents were interpreted with the chessboard system provided by the manufacturer to determine serotype or serogroup.
Molecular methods. DNA extraction
Pneumococcal isolates stored in BHI broth with 12% glycerol at -80°C were subcultured on CBA in 5% CO2. After overnight growth, a loopful of bacterial culture was suspended in 200 µL of sterile phosphate-buffered saline (PBS). A high pure PCR template preparation kit (Roche, Meylan, France) was used to extract DNA from the sample. Cell lysis and protein degradation were achieved by adding 10 mg/mL each of lysozyme and proteinase K, after which the temperature was increased to 70ºC. The DNA was then precipitated in isopropanol and recovered by centrifugation at 8000 rpm for 1 min. Finally, following several washing steps, the DNA was eluted in 100 µL of elution buffer and stored at -80ºC for polymerase chain reaction (PCR) analysis.
Molecular capsular multiplex PCR serotyping
Gene-based serotyping was performed using multiplex sequential PCR, as described by Pai et al.20 Briefly, PCR was carried out by first grouping 29 primer pairs into 7 multiplex PCRs targeting different serotypes. A positive control consisting of S. pneumoniae ATCC 49619 was included in all PCRs, in addition to an internal control targeting the cpsA gene found in pneumococci. Multiplex PCRs were performed using a multiplex PCR kit (Qiagen, Mainz, Germany), and each of the primers was used at equal final concentrations of 0.2 µM. Amplification was carried out with the following conditions: an initial denaturation step at 95ºC for 15 min, followed by 30 cycles of repeated annealing (94ºC for 30 s), polymerization (54ºC for 90 s), and extension (72ºC for 90 s). All PCR products were examined by 2% agarose gel electrophoresis.
Detection of resistance genes
Streptococcus pneumoniae isolates were subjected to multiplex PCR to detect the resistance genes pbp2x, pbp2b, and pbp1a (which confer resistance to ß-lactams), as well as mefA and ermB (which confer resistance to macrolides, lincosamides, and streptogramins).15,20,21 The 6 primer pairs were all included in one multiplex PCR. A primer pair for the specific marker gene of pneumococci (cpsA) was included in each PCR as an internal control. One positive control consisting of ATCC 49619 (cpsA+, pbp1a+) was included in each run, along with a negative control (molecular grade water). The multiplex PCR reactions were performed using a multiplex PCR kit (Qiagen, Mainz, Germany). All primers were used at equal final concentrations of 0.2 µM. The cycling conditions were tested and set as follows: an initial denaturation step at 95ºC for 15 min, followed by 40 cycles of repeated annealing (94ºC for 30 s), polymerization (60ºC for 90 s), and extension (72ºC for 60 s), with a final extension at 72ºC for 10 min. All PCR products were examined by 2% agarose gel electrophoresis.
Statistical analysis
Fisher’s exact test was conducted using the GraphPad Prism version 6.0 for Windows to compare the geno/serotype frequencies between various susceptibility groups of bacterial isolates. An analysis of variance (ANOVA) was performed using the same software to test for correlations between patient age and serotype prevalence. A p-value of ≤0.05 denoted statistically significant differences among different strains.
Results
Among the 94 tested isolates, 77 isolates were confirmed as S. pneumoniae at the phenotypic and genotypic levels, while the other 17 did not express the housekeeping gene cpsA (cpsA-, mean age: 28 years) (Figure 1, Tables 1 & 2). Twenty-four isolates did not belong to any of the serotypes tested in our protocol and were labeled as non-typeable pneumococci (cpsA+, mean age: 9 years). In total, 11 capsular serotypes were identified and the results of phenotypic and genotypic analysis were 100% concordant (Table 1). Six serotypes dominated, representing 64% of the tested isolates: 11A (22.6%), 19A (15%), 17F (13.2%), 23F (11.3%), 3 (9.4%), and 19F (7.5%).
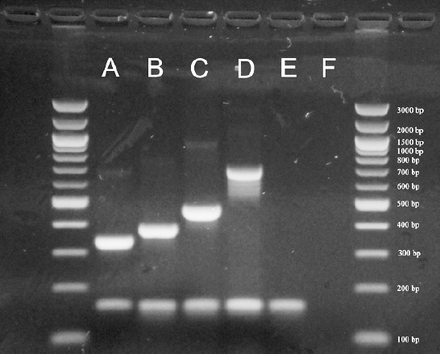
Example of serotyping results obtained by multiplex polymerase chain reaction (PCR). A= 33F (338 bp), B= 23F (384 bp), C= 11A/D (463 bp), D= 17F (693 bp), E= Streptococcus pneumoniae ATCC 49619 (cpsA+), 160 bp), F= negative control.
Different pneumococcal serotypes detected along with the mean age of patients.
Tests results for antimicrobial resistance to ß-lactams and macrolides.
Antimicrobial resistance
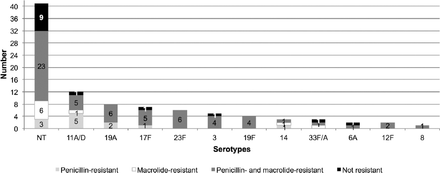
This study investigated pneumococcal antimicrobial resistance to ß-lactams and macrolides (Figure 2). Among the 53 typeable isolates, 48 (90.6%) were shown to be antimicrobial-resistant and only 5 isolates (9.4%) were non-resistant (Table 2). Non-typeable isolates (cpsA+ and cpsA-) were also tested for resistance to ß-lactams and macrolides (Table 2). The results showed that the majority (62.5% and 47%) was resistant to both penicillin and macrolide, and 4 and 5 isolates, respectively, were nonresistant. The distribution of the antimicrobial resistance patterns within the identified serotypes is presented in Figure 3. The most prevalent serotype, 11A/D, showed the highest resistance to penicillin of all the identified serotypes (p=0.76). Overall, the results showed high resistance to both penicillin and macrolides (67.9%) for the majority of serotypes, followed by penicillin only (17%) and macrolides only (5.6%).
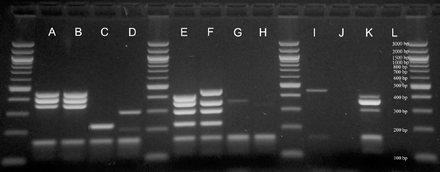
Example of resistance gene patterns results obtained by multiplex sequential PCR. A, B - resistant to penicillin, C, D - resistant to macrolides, E, F - resistant to penicillin and macrolides, G, H - not resistant, I, J - cpsA-, K - S. pneumoniae ATCC 49619 (cpsA+, pbp1a+), L - negative control.
Number of non-typeable (NT) and typeable isolates resistant to penicillin, to macrolides, to both penicillin and macrolides, and non-resistant. P-value as calculated by Fisher’s exact test was 0.76, suggesting that resistance patterns did not significantly correlate with serotype.
Two genes were studied to investigate resistance to macrolides (ermB and mefA). Results showed 39/48 (81.6%) of S. penumoniae typeable isolates were resistant to macrolides. Genotypes ermB+ were found in 2 (4.2%), mefA+ in 14 (29.2%), and ermB+ mefA+ in 23 (47.9%) of the resistant isolates, respectively (Table 3). Non-typeable isolates (cpsA+ and cpsA-) were also studied and the corresponding resistance genes were detected as follows: mefA+ (cpsA+: 9 (37.5%), cpsA-: 5 (29.4%), ermB+ mefA+ (cpsA+: 7 (29.2%), cpsA-: 3 (17.6%), and ermB+ (cpsA+: 3 (12.5%), cpsA-: 2 (11.8%).
Distribution of macrolide resistance-encoding genes in the detected pneumococcal serotypes (typeable, cpsA+) and non-typeable isolates (cpsA+ and cpsA-).
Discussion
This work was undertaken to study the relative prevalence of respiratory S. pneumoniae serotypes after routine use of a 13-valent pneumococcal conjugate vaccine (PCV13) and their patterns of antibiotic resistance. The main limitation of this study was the low yield of conventional culture in pneumococcal disease and the possible underestimation of a subset of pneumococcal respiratory disease.
The most prevalent serotypes found were 11A/D, 19A, 17F, 23F, 3, and 19F, in decreasing order. The most prevalent serotypes, 11A and 19A, have been shown to cause increased mortality in a recent meta-analysis.22 Although current data suggests that the 11A serotype can be carried in the nasopharynx of children in a non-invasive status, there is evidence that it can act as a precursor for various pathogenic serotypes in vitro. This serotype switching can be driven by unstable genetic regions such as tandem repeats.23 In addition, 11A causes a high fatality rate in adults due to pneumonia complicated by sepsis.24 The clinical relevance of the infection in a young adult age group (mean age, 29 years; p=0.001) by this serotype seen in this cohort (Table 1) requires further investigation in larger multicenter surveillance programs, with the knowledge that this capsular type is covered by the available pneumococcal polysaccharide vaccine (PPV23).7 Previous epidemiological studies of serotype 11A have led to the discovery of the new serotype, 11E, which is identical to serotype 11A except that it possesses an inactivated wcjE gene and is not bound by ficolin-2. This attribute is thought to be linked with its ability to bind to ficolin-2, fostering opsonization by phagocytes in the host.25 In a neighboring country, Iran, 11A/D was reported as a rare serotype (2.5%).26 Comparable studies in the Arabian Gulf, Middle East, and North Africa region (MENA) have revealed 19F, 3, 19A and 23F to be among the most common serotypes, similar to our findings.27 Soto-Noguerón et al28 studied the association between S. pneumoniae serotypes and mortality and found that 2 of the serotypes identified in our study (3 and 19A) were significantly associated with poor prognosis in both adult and pediatric age groups.
Conjugate vaccines usually include capsular pneumococcal serotypes that help protect against nasopharyngeal carriage. Previous studies in the United States have suggested that the incidence of serotypes included in the heptavalent vaccine (PCV7) has dramatically decreased to near zero, due to the vaccine’s wide use; other serotypes not included in the vaccine have remained stable or increased, with evidence of variable cross-reactivity.24,29 The PCV7 vaccine was first introduced in this country in 2006; initially, it was introduced locally on a limited scale, then as a part the National Immunization Program in 2008, before the switch to PCV13 in 2010. Despite the use of the PCV7 vaccine in Saudi Arabia, we found in this study that some of the PCV7 serotypes (19F, 23F, 14, 6B) are still prevalent in the country. Moreover, serotypes 3, 6A, and 19A, which are covered in PCV13, still constitute 29% of the pneumococcal burden in respiratory diseases (Table 1). Al-Sherikh et al18 investigated the distribution of pneumococcal serotypes in Riyadh, and found vaccines PCV7 to provide (77%), PCV10 (81%), and PCV13 (90%) serotype coverage.18 On the other hand, a study of invasive pneumococcal disease (IPD) among Saudi children aged less than five years (between 2005 and 2010) showed these three vaccines to offer 53%, 80%, and 91% serotype coverage, respectively.10 In Lebanon, a 6-year study including 78 hospitals showed 51%, 74% and 80% of PCV7, PCV10 and PCV13 coverage respectively for children of 2-5 years of age.27 A recent review done in Saudi Arabia stated that different IPD causing pneumococcal serotypes vary between the different geographic areas and the vaccination periods of the studies.7 Despite the diverse demographic studies listed above and the different vaccines coverage obtained, the majority concluded on the urgent need of current monitoring of prevalent pneumococcal serotypes in order to evaluate current vaccines impact on IPD epidemiology.
Serotyping results showed a 100% match between phenotypic (Pneumotest-Latex) and genotypic assays, demonstrating the high sensitivity of the sequential multiplex PCR method, therefore rendering cumbersome conventional methods unnecessary, especially in institutions where the infrastructure for molecular assays is well established. Yet there were some discrepancies between phenotypic and genotypic identification with regard to the presence of a capsule. Molecular testing found a high percentage of isolates (43.5%) to be non-typeable, of which 25.5% were lacking the capsule gene cpsA. In most serotypes, the capsule operon is flanked by the genes dexB and aliA. The first 4 capsule genes, cpsABCD, are conserved, and serotype-specific genes are located downstream of these. CpsA appears to act as a transcriptional activator of polysaccharide production but is not necessary for encapsulation.24 Unencapsulated pneumococcal strains evolve through either downregulation of the capsule or loss of the capsular biosynthesis locus.30 Non-typeable isolates of S. pneumoniae have been reported increasingly following vaccination and one earlier study found them to represent 28% of pneumococcal isolates, although the sample size of pneumococcal isolates was small (59 isolates) similar to that of our work.31 Similarly, Selva et al31 found that 24% of 98 pneumococcal isolates lacked a cpsA gene in direct testing of pleural samples, a useful rapid tool for pneumococcal detection in cases of complicated community-acquired pneumonia.32 The plasticity of the S. pneumoniae genome allows for serotype conversions when a missense mutation inactivates enzymatic activity of the involved gene, which, along with the high degree of homology in the sequences of capsule regions, makes it challenging to design primers for more serotypes.33 This may explain the apparent rise of non-typeable pneumococcal strains.
Dixit et al have reported increases in capsular serotypes not covered by the current vaccines and in unencapsulated S. pneumoniae after the introduction of PCV and PPV.34 Our results reinforce these observations and raise concerns about the possible emergence of new pathogenic pneumococcal strains not covered by the vaccines currently available. These infectious agents of invasive diseases require extensive study focusing on their phenotypic and genotypic features for future classification and clinical control. On the other hand, the World Health Organization (WHO) discussed pneumococcal vaccines efficacy. According to their report, different factors (such as geography, time and surveillance methods) are leading to the observation of vaccines uncovered serotypes emergence, including but not limited to the use of pneumococcal vaccines.34 Moreover and conferring to the same report, a high quality surveillance of IPD should be a population-based study starting 2 years before PCV introduction and lasting at least 5 years post- PCV introduction. S. pneumoniae strains were also studied for the occurrence of genes conferring resistance to penicillin and macrolides. The results showed high overall rates of resistance to both classes (67.9%) in both typeable and non-typeable isolates (62.5% of cpsA+ and 47% of cpsA- isolates). This percentage is high compared with that found in a prospective 6-year study in Saudi Arabia and Lebanon.27,33 Nine of our isolates, representing 4 serotypes (11A/D, 19A, 17F, and 14), were resistant only to penicillin, with the majority of isolates belonging to 11A/D, the predominating serotype in this cohort. Two macrolides resistance mechanisms were studied, focusing on the 2 resistance-encoding genes mefA and ermB. Contrary to the results found by Taha et al,36 we found that the gene mefA, responsible for efflux pump-mediated resistance, was more prevalent than ermB. A recent study conducted in Iraq also detected mefA at a higher rate than ermB.37
Detection of the erm gene mediating macrolide resistance in respiratory strains is of clinical concern. This gene encodes resistance to both older and newer generations of macrolides. Its extended spectrum also leads to inducible resistance to lincosamides such as clindamycin, an agent used to treat severe community-acquired pneumonia, and streptogramin agents, which are reserved for treating extensively drug-resistant (XDR) bacterial infections.38,39 All 6 of the isolates of 23F serotypes exhibited this gene (Table 3). This finding is supported by a previous report of a similar clone linked to an international pneumococcal outbreak.40 Although less invasive than other serotypes, the 23F serotype has been shown to provoke a severe inflammatory response in an in vivo experimental model of pneumococcal meningitis and is suspected to be linked with increased virulence.41 Pneumococcal resistance to macrolides, unlike resistance to ß-lactams, is known to be absolute and cannot be overcome by increasing the dose of macrolides or related drugs even if the isolate exhibits a low minimal inhibitory concentration above the breakpoint.14 Thus, macrolide resistance in pneumococci represents a therapeutic challenge, especially when accompanied with resistance to ß-lactam drugs, the agents of choice for pneumococcal diseases.
In conclusion, our study described the post-vaccination distribution of serotypes among respiratory pneumococcal isolates in Eastern Saudi Arabia and the alarmingly high rates of antimicrobial resistance among them. Despite the study’s small size, it revealed that S. pneumoniae is still a major pathogen in several age groups, even in a high-income country with a framework that supports the implementation of routine immunization programs. Serotypes 23F and 19A are components of all commercially available pneumococcal vaccines, yet they still circulate in the community. These serotypes represent a therapeutic challenge, especially when accompanied with the genetic promiscuity of this respiratory pathogen through transformation. In particular, the emerging serotype 11A needs to be considered in the future development of conjugate vaccines. More importantly, a large subset of isolates was not typeable by current techniques, underlining the emergence of possible atypical pneumococcal serotypes not targeted by the currently available vaccines. Continued surveillance of pneumococcal serotypes is thus critical to the guidance of both national and international vaccine strategies. In addition, further characterization of the clonal dissemination of various strains will provide insight into their transmission.
Footnotes
Disclosure. Authors have no conflict of interest, and the work was not supported or funded by any drug company.
- Received November 28, 2016.
- Accepted February 15, 2017.
- Copyright: © Saudi Medical Journal
This is an open-access article distributed under the terms of the Creative Commons Attribution-Noncommercial-Share Alike 3.0 Unported, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.