Abstract
Objectives: To investigate the relationship of 3 single nucleotide polymorphism (SNP) variants of ADAM33 with asthma susceptibility in patients from Northern and Central Punjab, Punjab, Pakistan.
Methods: In this case-control study, healthy and asthmatic participants were recruited between 2015 and 2017. The SNPs of ADAM33 gene, rs2280089, rs2280090, and rs2280091 were analyzed in 296 asthma patients and 343 healthy controls, as well as linkage disequilibrium and haplotype analysis.
Results: The non-significant differences were observed in allele and genotype frequencies of the SNPs in asthmatic and healthy persons even after population stratification based on age, caste, gender, family history, and environment. Although these SNPs were non-significant for disease susceptibility among children and adults, a fixed unique pattern of inheritance was nevertheless observed for the studied SNPs. Linkage disequilibrium analysis presented a very strong linkage between the SNP variants to predict their co-inheritance in study population. However, none of the haplotypes were found to be associated with asthma disease development.
Conclusion: The studied SNPs of ADAM33 appeared to be non-significant for asthma susceptibility in Northern and Central Punjabi population. The fixed allele combination inheritance pattern was a unique observation contrary to findings in other global populations.
Asthma is a chronic respiratory disorder characterized by inflammation and excessive mucosal secretion leading to airflow restriction and bronchial hyper responsiveness, consequently resulting in irreversible structural changes and airway remodeling. The prevalence of asthma is increasing globally, and it is anticipated that asthma, along with chronic obstructive pulmonary disease (COPD), may become the third leading cause of global mortality in the near future.1,2 The prevalence of allergic and respiratory disorders has increased substantially in Pakistan, and the current percentage of physician-diagnosed asthma patients is about 9.5%; however, this number is continuously increasing in line with trends in other Asian countries.3,4
Asthma is a polygenic disease triggered by a complex interplay between environmental and genetic factors, leading to diverse phenotypes with variable severity and atopic reactions.5-7 Increased familial aggregation of the disease, as shown by the observed increase in concordance in monozygotic twins relative to that in dizygotic twins, is an indicator of major genetic/genomic influence on asthma disease development.8 More than 100 genomic loci have been reported to be linked with asthma disease susceptibility;2 among them ADAM33 is a major asthma susceptible candidate gene.9
The ADAM33 gene was first reported in an outbred population as an asthma manifestation-associated gene.10 It contains 23 exons and is extended over a 14-kb region of chr. 20p13. It encodes the membrane-anchored enzyme disintegrin and metalloproteinase domain-containing protein 33, which influences the activity of growth factors and cytokines that are involved in the remodeling of airways.10,11 ADAM33 has been extensively studied as an asthma risk factor because of its likely role in cell differentiation, abnormal proliferation, and airway remodeling.12,13 The single nucleotide polymorphisms (SNPs) of ADAM33 gene have been studied in subjects of various ethnicities, but the strength of the association of these variants with asthma development has been found to vary in different populations, and several studies have reported contradictory results regarding the association of these SNPs within sub-ethnic groups of the same population.3,8
In previous studies, the rs2280089, rs2280090, and rs2280091 SNP variants have shown significant as well as non-significant associations with asthma susceptibility in different asthmatic populations in Asia.14,15 Owing to the conflicting and variable results regarding the impact of these SNP variants of ADAM33 in manifestation of asthma,8,16 the current study was designed to determine the probable role of 3 SNP variants [rs2280089, rs2280090, and rs2280091] of ADAM33 in the pathogenesis of asthmatic complications in a local population of Northern and Central Punjab, Punjab, Pakistan.
Methods
A total of 296 clinically diagnosed asthma patients and 343 normal healthy participants from Northern and Central Punjab (Table 1) were enrolled for this study between 2015 and 2017. The asthma patients were selected from Asthma and Allergy Clinics for collecting peripheral blood samples after being diagnosed by pulmonologists. Healthy control participants without any history of allergy, atopy, asthma, or other severe diseases, were randomly recruited from the general population for this case-control association study.
Demographics data of study participants.
This study was approved by the Ethical/Institutional Review Board of The Children’s Hospital & Institute of Child Health, Lahore and the University of the Punjab, Lahore, Pakistan. Signed informed consent, written in the local language, was obtained from recruited study participants or guardians in case of minors and all experiments were in accordance with the Helsinki Declaration.
Asthmatic patients with a history of any sickness involving the airway/lungs, such as emphysema, chronic bronchitis, pneumonia, tuberculosis, and so forth were excluded from this study. A detailed history was recorded for the non-asthma controls to exclude participants with any history of pulmonary disorders, allergies, or a family history of such diseases.
Single nucleotide polymorphism detection strategies
In current case-control study, target SNP variants were studied principally by candidate gene approach. Different strategies are generally adapted to study SNP variants in candidate gene approach based upon availability of financial resources and research instruments. Restriction Fragment Length Polymorphism (RFLP), allele specific primers, real time polymerase chain reaction (PCR), SNaPshot (single base extention method) and DNA sequencing are most popular strategies to study SNP variants in candidate gene approach.8,15,17,18 However we have adapted DNA sequencing strategy which is little bit expensive than other techniques but it is most reliable technique for detection of SNPs.
Deoxyribonucleic acid extraction and sequencing
Genomic DNA was extracted from blood samples using a previously described organic extraction method.19 Target SNPs of ADAM33 were analyzed using capillary-based sequencing on GA-3130XL (Applied Biosystems, Foster City, CA, USA). Single nucleotide polymorphisms regions were first amplified by PCR in a 20 µL reaction volume containing 20 ng DNA, 1 µL primer pair (8 µM each), and 10 µL 2x ready reaction PCR mix (EZ D-PCR MASTER MIX, Bio Basic, Canada). Amplified PCR products were purified by HighPrep PCR Clean Up kit (MagBio Genomics, USA), and the purified PCR amplicons were used as templates for sequencing PCR. Big Dye kit ver. 3.1 (Thermo Fisher Scientific, USA) was used for sequencing, and the products were further analyzed on GA-3130XL (Applied Biosystems, USA). Sequencing results were analyzed and interpreted by Sequencing Analysis 5.1 software (Applied Biosystems) (Table 2).
Amplification primer sequences.
Statistical analysis
SHEsis software, a powerful platform used for the analysis of linkage disequilibrium (LD), haplotype construction and genetic associations with polymorphic genomic loci,20 was used to analyze the allele frequencies, genotype frequencies, LD, and to perform haplotype construction. Haploview software version 4.2 was also used to verify the LD and haplotype results. The cutoff value was selected as p=0.05.
Results
Allele and genotype frequencies
Allele and genotype frequencies of participants are given in Table 3. No significant association was observed between the investigated SNPs and asthma in either the allelic or genotyping models. The association of these SNPs with asthma was also studied after performing population stratification. No association was observed at the sub-population level for Central or Northern Punjab. The association was also non-significant for study participants belonging to those groups, who were placed in category “others.” Similarly, no associations were observed after population stratification based on age, gender, family history of asthma, or urban/rural/semi-urban populations.
Allele and genotype frequencies of study participants.
Linkage disequilibrium analysis
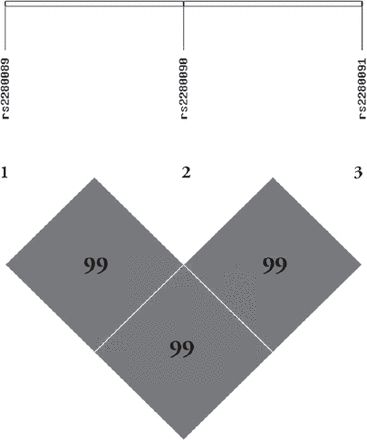
Linkage disequilibrium analysis revealed a very strong linkage between the studied SNPs in the population under investigation (Figure 1). There were no significant differences between the allele frequencies in either the asthmatic or control samples; for this reason, LD blocks were generated in a mixed population after excluding samples with missing genotype results (n=294 case, n=342 controls). Interestingly all 3 SNPs showed high LD (D¢≥0.994, r2 ≥0.983), even after population stratification. These SNPs followed a fixed inheritance pattern, as shown in Table 4.
Linkage disequilibrium (LD) block generated using Haploview software. Minimum genotyping call rate for all 3 single nucleotide polymorphisms (SNPs) was 90%. All samples, including cases and controls, were used for this analysis
Inheritance pattern in enrolled study participants.
Haplotypes
Haplotypes were generated in SHEsis software by considering 0.03 as lowest frequency threshold in haplotype analysis (Table 5).
Haplotype analyses.
Discussion
Different polymorphic variants of ADAM33 have been reported to show both a significant and non-significant association with asthma susceptibility in diverse populations. However, its potential genetic association with asthma cannot be ignored, because the investigated polymorphic variants may also be independently or collectively associated with the pathogenesis of asthma. However, the varying results in ethnically different populations suggest that association of polymorphic variants of ADAM33 with asthma differs according to ethnicity.12,15
In previous studies, the rs2280089, rs2280090, and rs2280091 SNP variants have shown significant as well as non-significant associations with asthma susceptibility.14 In the current case-control study, an analysis of these variants revealed that the minor allele and genotype frequencies showed no noticeable differences, indicating that the rs2280089, rs2280090, and rs2280091 markers may not be independently associated with asthma in the study population. Population stratification analysis demonstrated that there was no significant association for ethnic-specific participants in the Central Punjab, Northern Punjab, and “others”. The allele and genotype frequencies of these variants were also found to be non-significant after stratification based on age, gender, family history of asthma in asthmatics, and urban/rural/semi-urban population. Despite the reported significant associations of these SNPs with asthma in multiple populations, no significant association has been identified in sub-ethnic populations of the same origin.8,14,16,23 A recent study based on 2 cities of Pakistan (Islamabad and Lahore) reported a protective role of rs2280091 against asthma in a Pakistani population.24 As the study was based in Islamabad city, the capital of Pakistan, the participants included an admixture of different ethnicities within Pakistan. It is highly probable that the major component of the study population originated from Islamabad (not described in that study), which may be the reason why our results differed from those in the previous study. However, ethnicity-based studies in other provinces of Pakistan are required to improve our understanding of the role of the studied SNPs in the development of asthma.
The observed non-significant association with asthma in a Northern and Central Punjabi population was consistent with the results of our previous study, which was specific to the Lahore region.25
Linkage disequilibrium analysis is used to determine the combination of alleles at proximal genomic regions, which may predict non-random associations of investigated alleles at proximal loci.26 A very strong LD (D¢ >0.99) was observed in the SNPs studied here (rs2280089, rs2280090, rs2280091) in all subjects, suggesting that these SNPs are inherited together. Similarly, a strong LD (D¢ >0.99) was observed after population stratification based on age, gender, family history of asthma, urban/rural/semi-urban population, Northern Punjab, Central Punjab, and others. There was no observed difference in allele combinations in the studied participants, excluding a single healthy participant (ID= SGD-CNT-18, Female) who was recruited from Sargodha and belonged to the ethnically Awan family of Northern Punjab. All study participants, excluding SGD-CNT-18, followed the same allele inheritance pattern with 3 combinations: Pattern-1, GG GG AA; Pattern-2, AG AG AG; and Pattern-3, AA AA GG, for the 3 investigated SNPs (Table 4). This inheritance pattern has not been previously observed in a Northern and Central Punjabi population.
Qu et al27 investigated the same 3 SNPs in a Northern Chinese population, and reported that rs2280089 and rs2280091 were significantly associated with asthma, whereas rs2280090 was not significantly associated with asthma in a pediatric population. However, a strong LD among these SNPs was not observed, contrary to the findings in our population. Another recent study on a Chinese Li23 population reported the significant association of rs2280089, and the non-significant association of rs2280091 with susceptibility to asthma. The same study also predicted a low LD between the SNP variants in the Li population. These findings of a low LD for the target SNPs were also reported in others populations, including Mongolians and Egyptians.28,29
A Saudi Arabian population-based study previously reported a significant association between the rs2280091 and rs2280090 SNP variants and asthma susceptibility, which is contradictory to the current findings; however, that study also reported a strong LD between the 2 variants, consistent with the findings of our study.30 A recent study on a Southwestern Iranian population also reported similar results to those of the current study regarding the non-significant association of rs2280089 and rs2280091 with asthma disease, as well as a strong LD between these SNPs.8 Zeinaly et al also have reported the non-significant association of rs2280091 SNP with asthma in Azerbaijan Population of Iran.15
Study limitations
Punjab province of Pakistan is an admixture of populations that is culturally similar but even though ethnically/linguistically and genetically different races/tribes live in different regions of Pakistani-Punjab. Our study mainly covered Northern and Central Punjab with limited sample size; therefore, studies with increased sample size are suggested from different regions of Punjab which may help in better understanding the role of ADAM33 gene’s SNP variants against asthma susceptibility.
In conclusion, we did not find any significant association of the rs2280089, rs2280090 and rs2280091 SNPs with susceptibility to asthma in allelic or genotypic models in this Central and Northern Punjabi population. Haplotype analyses results were also nonsignificant for these SNPs in terms of asthma susceptibility. The findings of this study report a very strong LD among the studied SNPs, along with a statistically non-significant association with asthma disease in Northern and Central Punjab. These findings support the hypothesis that genetic association of ADAM33 gene’s polymorphic variants against asthma susceptibility is ethnicity-specific. Further similar studies may enhance our ethnicity-based knowledge regarding asthma progression in order to develop customized approaches for more accurate diagnosis and prognosis in ethnically varied populations.
Clinical Practice Guidelines
Clinical Practice Guidelines must include a short abstract. There should be an Introduction section addressing the objective in producing the guideline, what the guideline is about and who will benefit from the guideline. It should describe the population, conditions, health care setting and clinical management/diagnostic test. Authors should adequately describe the methods used to collect and analyze evidence, recommendations and validation. If it is adapted, authors should include the source, how, and why it is adapted? The guidelines should include not more than 50 references, 2-4 illustrations/tables, and an algorithm.
Acknowledgment
We would like to thank Editage (www.editage.com) for English language editing.
Footnotes
Disclosure. Authors have no conflict of interests, and the work was not supported or funded by any drug company.
- Received May 2, 2019.
- Accepted July 16, 2019.
- Copyright: © Saudi Medical Journal
This is an open-access article distributed under the terms of the Creative Commons Attribution-Noncommercial-Share Alike 3.0 Unported, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.