Abstract
Objectives: To investigate the role of cell cycle protein-dependent kinase regulatory subunit 1B (CKS1B) in driving the aggressive and rapid proliferation observed in pancreatic cancer.
Methods: A comprehensive analysis was carried out using raw mRNA information and data from 2 databases: the cancer genome atlas and gene expression omnibus. The differential expression of CKS1B at the mRNA and tissue levels in cancer and adjacent paracancerous tissues were assessed. Additionally, the relationship of CKS1B expression and overall survival (OS) rate was investigated using Kaplan-Meier survival curves. Potential molecular mechanisms by which CKS1B may influence the biological characteristics of pancreatic cancer were explored using resources available within the encyclopedia of RNA interactomes database.
Results: The CKS1B exhibited significant differential expression at the mRNA as well as protein levels. A correlation with statistical significance between CKS1B expression and N stage, age, and alcohol consumption was observed. Notably, high CKS1B expression was determined as a predictive factor for worse OS. Furthermore, the analysis revealed a potential synergistic role between CKS1B and the molecule PKMYT1, which could impact the ATR-Chk1-Cdc25 signaling pathway and disrupt the G2/M checkpoint within the cell cycle, ultimately promoting abnormal tumor proliferation.
Conclusion: The CKS1B may serve as a novel potential prognostic factor in pancreatic cancer and is involved in the abnormal proliferation biology phenotype by mediating cell cycle signaling pathways.
Pancreatic cancer poses a formidable challenge in the field of oncology, presenting a serious threat to human life and well-being. The latest American cancer statistics for 2022 reveal that pancreatic cancer stands as the fifth leading cause of mortality in the realm of malignancies.1 However, the current therapeutic options offer limited cause for optimism. Traditional treatment approaches, such as surgery, are only viable for a minority of pancreatic cancer patients. Despite remarkable advancements in clinical research pertaining to radiation therapy, chemotherapy, and immunotherapy, the long-term survival rate remains disappointingly stagnant.2 Consequently, the quest for novel prognostic markers within the context of pancreatic cancer assumes heightened significance. It is imperative to explore innovative approaches that could potentially yield more efficacious interventions and improved prognoses for individuals afflicted by this formidable malignancy.
The utilization of tumor-based bioinformatics is crucial for uncovering and predicting molecules associated with tumor prognosis. In this study, bioinformatics was adopted to screen and analyze multiple tumor databases, specifically focusing on identifying differential genes in pancreatic cancer. This investigation led to the identification of regulatory subunit 1B (CKS1B), a potential target molecule that exerts vital effects on regulating cell cycles.3,4 The CKS1B exhibits abnormal expression patterns in various tumor tissues, including non-small cell lung as well as gastric cancers, and participates in driving biological processes like abnormal proliferation, invasive metastasis, as well as chemotherapy resistance. This evidence highlights the significance of CKS1B as a pivotal gene in the progression of multiple tumors. Despite of that, there is a lack of comprehensive reports exploring the detailed mechanisms through which CKS1B influences pancreatic cancer development, presenting an essential and unexplored area for further investigation.
In this research, a suite of bioinformatics methods was employed to investigate CKS1B’s role in pancreatic cancer. These methods included differential gene expression analysis, validation of expression across various databases, and correlation analysis of CKS1B with critical clinical factors such as clinical stage and overall survival (OS) in pancreatic cancer patients. This study revealed CKS1B as a potential marker in the prognosis of pancreatic cancer. The CKS1B’s differential expression levels in tumor tissues as well as the paraneoplastic tissues were thoroughly assessed, identifying a significant correlation between heightened CKS1B expression and diminished OS among pancreatic cancer patients. Additionally, the plausible molecular mechanisms were explored, including the identification of upstream miRNAs that negatively regulate CKS1B and the delineation of its collaborating genes. These investigations shed light on the intricate mechanisms through which CKS1B influences the biological processes underlying pancreatic cancer.
Methods
In this study, separate analyses of mRNA differential genes were carried out using data obtained from 2 distinct databases: the cancer genome atlas (TCGA) and gene expression omnibus (GEO). Differential gene identification was carried out using the R programming language, applying specific criteria (p<0.05 and Log2 of >1 fold change). The set of genes identified as differentially expressed in both databases were collected, resulting in a collection referred to as the differentially expressed genes (DEGs), representing the intersection of genes meeting the established criteria in both datasets.
To validate the DEGs, including CKS1B, at the mRNA and protein levels, data from the TCGA and GEO databases were utilized for mRNA validation, while the human protein family tree database was employed for protein-level validation. Furthermore, the University of Alabama at Birmingham cancer data analysis portal (UALCAN) platform was utilized to explore the potential association between differentially expressed CKS1B and clinical phenotypes in pancreatic cancer, including tumour, node, and metastasis stage, age, and OS.
The LinkedOmics database (Texas, USA) was employed to explore the mechanism of action of differential genes. To predict molecules interacting with CKS1B, the search tool for the retrieval of interacting genes/proteins (STRING) database (Baden-Wuerttemberg, Germany) was utilized. To uncover potential biological functions of CKS1B and its interacting molecules, data from the gene ontology (GO, Maryland, USA) as well as Kyoto encyclopedia of genes and genomes (KEGG, Tokyo, Japan) databases were referenced. Additionally, the encyclopedia of RNA interactomes (ENCORI) database (Guangzhou, China) was used to identify microRNAs (miRNAs) that may regulate CKS1B and influence its prognosis in pancreatic cancer.
Statistical analysis
The “Limma” R package was utilized to detect differential genes, implementing paired t-tests with a cut-off set at p<0.05. The authors used GraphPad Prism version 8.4.3 (California, USA) for the analysis of CKS1B’s expression among cancerous tissues as well as normal tissues of gastric cancer in TCGA and GEO databases by Wilcoxon t-test, to find out the differential expression between the groups. The UALCAN platform was adopted for comparing CKS1B’s differences with the different clinical grades and classifications based on the ANOVA analysis test. The LinkedOmics database was used to screen for CKS1B-related genes using Pearson’s correlation analysis. A p-value of <0.05 was considered significant.
Results
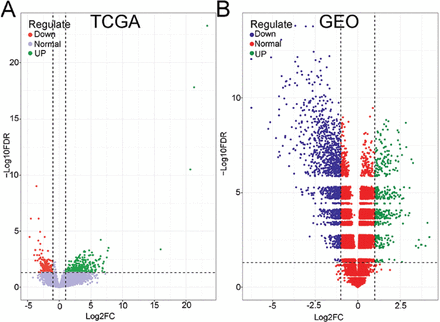
This study analyzed 178 pancreatic cancer tissues and 4 paraneoplastic tissues from TCGA, as well as 36 pancreatic cancer tissues and 16 normal tissues from the GEO database. The goal was to compare gene expression levels between these datasets, and the findings were visually represented through volcano plots. In the TCGA dataset, a total of 3,586 differential genes were identified, including 2,410 upregulated genes and 1,176 downregulated genes (Figure 1A). In the GEO dataset, 1,874 differential genes were discovered, with 1,372 upregulated genes and 502 downregulated genes (Figure 1B). There were 576 common differential genes found between these 2 databases, consisting of 423 upregulated genes and 153 downregulated genes. Among the 423 upregulated genes, CKS1B was particularly observed.
- Identification of differential genes. A) TCGA pancreatic cancer and paraneoplastic tissues differential genes. B) GEO pancreatic and paraneoplastic tissues differential genes. Green dots indicate a p-value of<0.05 and log2(FC) of >1, while red dots indicate a p-value of <0.05 and log2(FC) of <1. TCGA: the cancer genome atlas, GEO: gene expression omnibus, FC: fold change, FDR: false discovery rate
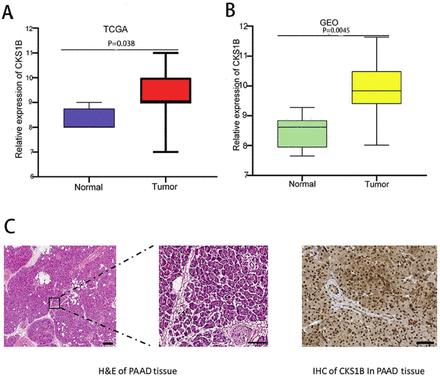
The CKS1B expression was increased in pancreatic cancer with statistical significance in contrast with normal neighboring tissues in both the TCGA and GEO databases (Figures 2A & 2B). To validate these results, CKS1B expression status at the mRNA and protein levels in pancreas and paraneoplastic tissues was compared utilizing the human protein atlas (HPA) database (Sweden, Europe). Immunohistochemical staining indicated strong positive staining for CKS1B in pancreatic cancer tissues, while it was negative in normal tissues (Figure 2C). These findings firmly establish CKS1B’s distinctive differential expression in pancreatic cancer.
- Differential expression of CKS1B. A) CKS1B expression in pancreatic cancer compared to normal and paraneoplastic tissues in TCGA. B) CKS1B expression in pancreatic cancer compared to normal and paraneoplastic tissues in GEO. In contrast with normal tissues, cancer tissues showed higher CKS1B expression with statistical significance in TCGA (p<0.05) and GEO (p<0.0001). C) Immunohistochemistry of CKS1B in normal and tumor tissues in the the human protein atlas database. Pancreatic cancer tissues on the left and normal tissues on the right. The CKS1B showed strong positive staining results in pancreatic cancer tissues while negative in normal tissues. CKS1B: cell cycle protein-dependent kinase regulatory subunit 1B, TCGA: the cancer genome atlas, GEO: gene expression omnibus, H&E: hematoxylin and eosin, PAAD: pancreatic adenocarcinoma, IHC: immunohistochemistry
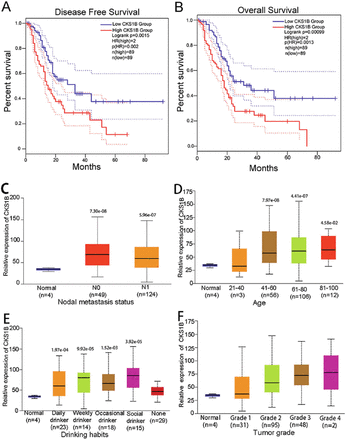
To understand the prognosis of pancreatic cancer patients, OS together with disease-free survival (DFS) were assessed. The gene expression pofiling interactive analysis (GEPIA, Beijing, China) database was used to investigate potential connections between CKS1B expression and OS as well as DFS in individuals with pancreatic cancer. Kaplan-Meier curves consistently demonstrated shorter survival for the high CKS1B expression group, but longer survival for the low CKS1B expression group. This trend was also observed in the DFS analysis (Figures 3A & 3B).
- Clinical phenotype analysis and survival of CKS1B. A) Progression-free survival (DFS) of CKS1B expression in the GEPIA database; high CKS1B expression group (red line) was significantly lower than low CKS1B expression group (blue line); p<0.05. B) Overall survival (OS) based on K-M curve of CKS1B expression; high CKS1B expression group (red line) was lower than that of low CKS1B expression group with statistical significance (blue line); p<0.05. C-F) Relationship between CKS1B expression and clinical phenotypes such as tumor stage, lymph node metastasis, age, and alcohol consumption in the University of Alabama at Birmingham Cancer data analysis portal database. CKS1B: cell cycle protein-dependent kinase regulatory subunit 1B
After confirming the differential expression of CKS1B among both malignancy and paraneoplastic tissues, the association of the elevated CKS1B expression with clinical phenotypes in pancreatic cancer was investigated. Genetic sequencing data from pancreatic cancer patients available in the UALCAN database, along with clinical follow-up information, were analyzed. Significant associations were found between CKS1B expression and specific clinical phenotypes. Notably, higher CKS1B expression levels were linked to lymph node staging and lymph node recurrence in pancreatic cancer (Figure 3C). Additionally, pancreatic cancer patients aged 40 years and older and those reporting alcohol consumption exhibited higher levels of CKS1B expression (Figure 3D & 3E). However, no substantial correlation was found between CKS1B expression and disease grade (Figure 3F).
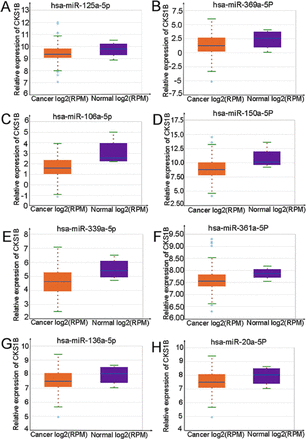
Given CKS1B’s pivotal impact on the prognosis of pancreatic cancer, researchers embarked on an investigation into the regulatory mechanisms of CKS1B within this particular malignancy. Initially, the complex mechanisms governing the expression of CKS1B were explored. The miRNAs, which are short, single-stranded non-coding RNAs, possess the remarkable ability to negatively regulate the mRNA levels of target genes, actively participating in the intricate processes of tumor development.5 With this knowledge in mind, researchers turned their attention to the ENCORI database, aiming to identify 78 upstream miRNAs that specifically target CKS1B. Among this vast array of miRNAs, 39 exhibited negative associations with CKS1B, while the remaining 39 displayed positive associations. Subsequently, the focus was narrowed down to the 39 miRNAs with negative associations, and a comprehensive analysis was meticulously carried out. This exhaustive analysis not only shed light on the regulatory network involving CKS1B but also uncovered 8 miRNAs that exhibited differential expression between malignant pancreatic tissues and their normal adjacent counterparts. Remarkably, Figure 4 shows that these 8 miRNAs (namely, hsa-miR-125a-5p, hsa-miR-361a-5p, hsa-miR-339a-5p, and more) possesses reduced expression levels in the cancerous tissues with statistical significance.
- Correlation analysis of CKS1B and miRNA. A-H) Differential expression status of hsa-miR-125a-5p, hsa-miR-369a-5p, has-miR-106a-5p, hsa-miR-150a-5P, hsa-miR-339a-5p, hsa-miR-361a-5p, hsa-miR-136a-5p, hsa-miR-20a-5P, as well as has-miR-106a-5p in the encyclopedia of RNA interactomes database versus normal tissues. CKS1B: cell cycle protein-dependent kinase regulatory subunit 1B
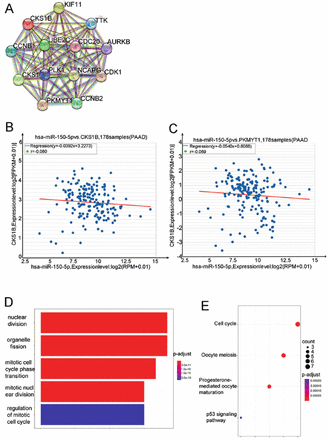
As a regulator of cell cycle proteins, CKS1B has been involved in tumor proliferation by influencing changes in the tumor cell cycle.5,6 Co-stimulatory molecules are often involved in this regulatory process. Therefore, an investigation into the co-stimulatory molecules associated with CKS1B was carried out. Genes exhibiting co-expression with CKS1B in the context of pancreatic cancer were identified utilizing the LinkedOmics database. Researches identified 218 genes with robust correlations (correlation coefficient of >0.6). Subsequently, the regulatory relationships of CKS1B with these co-expressed genes were investigated using the STRING database. This network of co-regulated genes included AURKB, CCNB1, KIF11, CCNB2, PLK1, UBE2C, TTK, AURKB, NCAPG, PLK1, and others (Figure 5A). Furthermore, an assessment was carried out to determine whether miRNAs acting upstream of CKS1B could influence these co-regulatory molecules. Comparative analysis using the ENCORI database revealed that out of the 10 potential co-regulatory molecules, only PKMYT1 served as a target gene regulated by hsa-miR-150-5P (Figure 5B & 5C).
- Regulatory relationship of genes co-expressed with CSK1B; A) connecting lines indicate the presence of regulation; the more connecting lines, the stronger the regulatory relationship. B&C) CKS1B enrichment analysis and co-expressed genes. Correlation analysis of PKMYT1 and CKS1B with hsa-miR-150-5P; D&E) the gene ontology as well as Kyoto encyclopedia of genes and genomes analysis of co-expressed genes in regulatory relationship with CKS1B. CKS1B: cell cycle protein-dependent kinase regulatory subunit 1B, PAAD: pancreatic adenocarcinoma
The GO analysis revealed the participation of CKS1B and PKMYT1 in critical biological processes, such as the mitotic cell cycle transition and overall cell cycle regulation. The KEGG analysis demonstrated their involvement in the ATR/Chk1/Cdc25 signaling pathway, which holds a pivotal role in regulating the cell cycle. Specifically, this pathway exerts a critical effect on governing the G2/M cell cycle checkpoint. Disruptions in this pathway can result in disruptions in the G2/M checkpoint, thereby leading to abnormal proliferation of tumor cells (Figure 5D & 5E).7,8
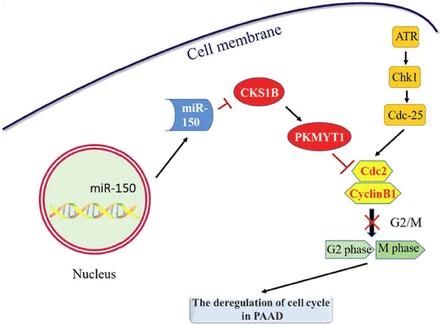
The molecular pathway by which CKS1B influenced pancreatic cancer involved downregulation of hsa-miR-150-5p, impacting the ATR-Chk1-Cdc25 signaling pathway. This disruption led to the upregulation of CKS1B, which in turn increased PKMYT1’s expression. The G2/M checkpoint’s malfunction in the cell cycle ultimately promoted abnormal proliferation of tumor cells (Figure 6).
- Signaling pathway analysis between CKS1B and PKMYT1; ATR-Chk1-Cdc25 signaling pathway involved in PKMYT1 during the cell cycle. CKS1B: cell cycle protein-dependent kinase regulatory subunit 1B, PAAD: pancreatic adenocarcinoma
Discussion
Pancreatic cancer is a highly malignant neoplasm with dismal prognostic outcomes, necessitating the expeditious discovery of novel therapeutic approaches to improve the 5-year survival rate.1 This research has carried out 3 noteworthy contributions.
Firstly, the findings of this study elucidated that CKS1B governs a distinct biological phenotype characterized by abnormal tumor proliferation, primarily stemming from the G2/M cell cycle checkpoint’s inactivation. The potential molecular mechanisms provide novel avenues for delving into the proliferation as well as the regulation of pancreatic cancer cells in future.
To elucidate CKS1B’s role, the authors initiated a comprehensive approach. The investigation commenced by examining CKS1B’s differential expression in pancreatic cancer and evaluating its prognostic implications using publicly available databases. Subsequently, the authors embarked on an exhaustive exploration of the upstream miRNAs of CKS1B and its downstream co-regulatory molecules. The CKS1B, belonging to the CKS/Suc1 protein family, is situated within the 1q21 region of the human chromosome. Its influence stems from its disruption of the cell cycle through interactions with and regulation of the catalytic subunit of cyclin-dependent protein kinases.9 Elevated CKS1B expression has been associated with heightened proliferation in breast cancer, colon cancer, and hepatocellular carcinoma cells (HCC), significantly impacting the prognosis of affected individuals, as documented in prior studies.10-12 Research has revealed that CKS1B, in conjunction with CKS2, plays a role in promoting HCC cell proliferation while simultaneously inhibiting chemotherapy-induced apoptosis.
Secondly, the authors identified a synergistic molecule, PKMYT1, which collaborates with CKS1B and unveiled a regulatory connection between PKMYT1 and Cdc2/cyclin B, offering novel insights that hold promise for potential treatments in the context of pancreatic cancer.
The CKS1B cooperates with CDK1 to expedite the transition of cancer cells from the G0 to the G1 phase, thereby contributing to their malignant proliferation.13,14 Hence, it is evident that CKS1B often operates in concert with synergistic molecules to regulate the cell cycle of tumor cells. In this study, one such synergistic molecule named PKMYT1, which collaborates with CKS1B, was identified, consistent with the aforementioned findings.
The PKMYT1, a serine/threonine protein kinase, is located on human chromosome 16 at the 16p13.3 locus. It functions as a bispecific protein kinase associated with the membrane, regulating the cell cycle via the phosphorylation of Thr14 as well as Tyr15. This action has implications for cellular structures such as the Golgi apparatus along with endoplasmic reticulum during the final stages of mitosis, where it inhibits the activity of key cell cycle proteins.15,16 Previous studies has emphasized the pivotal effect of elevated PKMYT1 expression on driving various malignancies, including ovarian cancer, gastric cancer, as well as colon cancer.17-19 The PKMYT1 has been shown to influence tumor growth by modulating Chk1/2 within the non-small cell lung-Cdc25C-Cdc2/cyclin B axis, inducing G2/M phase arrest.20 It has also been implicated in promoting the growth of prostate cancer cells through the regulation of CCNB1 in combination with CCNE1.21 Additionally, a PKMYT1 inhibitor, fostamatinib, has demonstrated efficacy in inhibiting the proliferation of prostate cancer cells. However, the relationship between PKMYT1 and Cdc2/cyclin B has been relatively underexplored, with limited prior studies. A regulatory connection through bioinformatics analysis was revealed, providing novel insights that hold promise for potential treatments in the context of pancreatic cancer.
Thirdly, the study culminated in the development of a comprehensive molecular signaling network, providing insights into the role of CKS1B in driving the aberrant proliferative phenotype observed in pancreatic cancer.
The miRNAs, small non-coding RNA oligonucleotides, play a pivotal role in gene regulation. Numerous miRNAs with deleterious regulatory functions are situated upstream of mRNAs and actively participate in the processes of tumorigenesis and tumor development.22-24 In this study, a specific miRNA, has-miR-150-5p, was identified through database analysis as a negative regulator of CKS1B. This miRNA has previously been linked to the advancement of nasopharyngeal and cervical cancers by suppressing PYCR1 and p27 Kip1’s expression. It is important to note that CKS1B can also be negatively regulated by other miRNAs.24 For example, miR-520h has been shown to inhibit CKS1B overexpression, mitigating the inhibitory effects on PACA-2 cells’ viability, migration, as well as invasion following LINC00657 knockdown. Furthermore, miR-1258 has been identified as another miRNA that negatively regulates CKS1B expression, leading to the inhibition of colorectal cancer cell proliferation, migration, and tumorigenicity.23,25,26 These findings shed light on the intricate regulatory network involving CKS1B and miRNAs in tumorigenesis.22 Among the miRNAs that regulate CKS1B upstream, it is noteworthy that only hsa-miR-150-5p possesses the unique capability to simultaneously regulate the co-molecule PKMYT1. This dual regulatory role significantly affects the G2/M checkpoint signaling pathway. The study culminated in the development of a comprehensive molecular signaling network, providing insights into the role of CKS1B in driving the aberrant proliferative phenotype observed in pancreatic cancer. This network is visually represented in Figure 6 for reference.
Study strengths & limitations
TBy employing a bioinformatic approach, the authors have identified CKS1B as a potential candidate involved in the abnormal proliferation and prognostic characteristics of pancreatic cancer. Although the study did not include in vivo or ex vivo molecular biology validations, the strength of the findings is rooted in the comprehensive data analysis carried out, the plausibility of the target gene, and the predictions regarding its upstream and downstream molecular network.
In conclusion, these findings hold significant promise for the treatment of pancreatic cancer. The CKS1B could offer fresh insights into both the treatment and prognosis prediction of pancreatic cancer. However, further research of the intricate molecular mechanisms underlying CKS1B’s role in cell cycle regulation and abnormal proliferation of pancreatic cancer is required in future.
Acknowledgment
The authors gratefully acknowledge CureEdit (www.wscureedit.com) for English language editing.
Footnotes
Disclosure. This study was supported by the Talent Introduction Project of prospective randomized controlled study of helical tomotherapy combined with chemotherapy versus chemotherapy alone for pancreatic cancer with oligo-metastasis, Beijing, China (no. first issue: 2022-2-5121 and 2021LC009).
- Received April 26, 2023.
- Accepted January 4, 2024.
- Copyright: © Saudi Medical Journal
This is an Open Access journal and articles published are distributed under the terms of the Creative Commons Attribution-NonCommercial License (CC BY-NC). Readers may copy, distribute, and display the work for non-commercial purposes with the proper citation of the original work.