Abstract
Objectives: To identify biomarkers that can discriminated small cell lung cancer (SCLC) from non-SCLC (NSCLC), and explore their association with the prognosis of SCLC under chemoradiotherapy.
Methods: The GSE40275 dataset was used to identify potential targets in SCLC. There were 196 patients of lung cancer (LC) in cohort 1 of this study. MTHFD1 levels in tissues were determined by immunohistochemistry assay in cohort 1. Lung cancer patients who were all underwent local chemoradiotherapy (CRT) were included in cohort 2, and the association of MTHFD1 levels with CRT treatment outcome were determined in cohort 2. Cell experiments were used to determine the function of MTHFD1 on the radio-sensitivity of SCLC and NSCLC cells.
Results: The MTHFD1 levels in LC tissues were increased, and could discriminate SCLC from both lung squamous cell carcinoma (LUSC) and lung adenocarcinoma (LUAD). Small cell lung cancer patients with MTHFD1 high phenotype had a poorer prognosis after CRT treatment, whereas no significant correlation was found between MTHFD1 levels and prognosis in LUSC and LUAD group. Cell experiments demonstrated that overexpression of MTHFD1 increases radio-resistance in both SCLC and NSCLC in vitro.
Conclusion: MTHFD1 expressions might be a novel specifically prognostic biomarker for SCLC and the CRT treatment outcome.
Lung cancer (LC) is not a single entity but rather a complex disease with distinct subtypes, primarily categorized into 2 major groups: non-small cell lung cancer (NSCLC) and small cell lung cancer (SCLC).1,2 Non-small cell lung cancer mainly includes lung squamous cell carcinoma (LUSC) and lung adenocarcinoma (LUAD). Generally, advanced age and positive smoking status, male gender, SCLC pattern, and advanced lung cancer stage (III or IV) are associated with poor prognosis.3,4
There are huge differences in molecular phenotypes between SCLC and NSCLC, and many studies have focused on identifying SCLC-specific genes and their biological functions to uncover the regulatory mechanisms and potential biomarkers for SCLC.5 Currently, the GSE40275 dataset, which is the RNA sequencing (RNA-seq) data determining the genes profiles of different subtypes of LC, has been uploaded to the GEO dataset. Through bioinformatics analysis of this dataset, we identified a series of SCLC related genes, including MTHFD1.
MTHFD1 is a gene that encodes a trifunctional enzyme involved in folate metabolism.6 Human MTHFD1 expressions and polymorphisms are reported to be closely associated with various malignancy.7-9
For SCLC and advanced NSCLC, a combined approach of radiotherapy and chemotherapy (chemoradiotherapy [CRT]) is the primary choice. However, there is a significant variation in the effectiveness of the CRT treatment. Here, we plan to investigate the relationship between MTHFD1 expression and the develop of different subtypes of LC, and their CRT treatment outcome.
Methods
This study is a prospective study, carried out from January 2020 to October 2023. The Eighth Medical Center of the Chinese PLA General Hospital, Beijing and Qingdao Municipal Hospital, Qingdao, China, were included in this study. The inclusion criteria were: I) histopathologically diagnosed LUAD, LUSC or SCLC; II) age 18-75 years; and III) complete clinical data. The exclusion criteria were: I) complicated with other malignant tumors; II) liver or kidney dysfunction; and III) pregnant or lactating women. A total of 196 LC patients, including 70 LUAD, 56 LUSC, and 70 SCLC cases were recruited as cohort 1, and their pathological tissues were used for subsequent detection. A total of 33 patients with inflammatory pseudotumor were included and their normal lung epithelial tissue samples were used as controls (HCs). Next, we selected patients who underwent radiotherapy and chemotherapy (excluding those who did not undergo radiotherapy and chemotherapy) in cohort1 to form cohort 2 (cohort 2 includes: 52 SCLCs, 30 LUADs, and 31 LUSCs). The study was carried out according to the Helsinki guidelines, with the approval from The ethics committee of the Eighth Medical Center of the Chinese PLA General Hospital, Beijing, China (approval ID: 3092023323152361). All participants provided their written consent.
Lung cancer tissues were homogenized, and then TRIzol™ LS Reagent (Thermo Fisher Scientific, Waltham, MA) were used to extract the total RNA from these tissues. Subsequently, the qRT-PCR detection was carried out by using PrimeScript RT-PCR kit (TaKaRa, Osaka, Japan) and SYBR Green PCR Kit (TaKaRa, Osaka, Japan) and MTHFD1 mRNA levels were determined by using the following primers:
Forward: 5’-GTTGAAGGAGCAAGTACCTGG-3’;
Reverse: 5’-GGTAGCTGCACTAAGAACCCA-3’;
Immunohistochemical (IHC) testing was carried out according to previous reports.7 The LC tissues or normal lung tissues samples were uniformly cut into 4 μm paraffin sections. Two experienced pathologists will independently interpret the IHC results. Scoring of the IHC will be based on the integrated optical density (IOD) measurement.7 Then, the expression of MTHFD1 will be uniformly normalized.
The human SCLC cell line H69, NSCLC cell line A549, and normal lung epithelial cell line BEAS-2B were purchased from the American Type Culture Collection (ATCC, USA). All cells were incubated in a humidified atmosphere containing 5% CO2 at 37°C with DMEM medium supplemented with 10% fetal bovine serum (GIBCO, USA).
For the radio-resistance assay, cells were instantly transfected with MTHFD1 over-expressed plasmid or control plasmid by using Lipofectamine2000 for 24 hours before being exposed to different doses of irradiation. After 48 hours post-irradiation, MTT assay was carried out using the commercialized MTT kit (Sigma, USA) in accordance with the instruction manual.
The GSE40275 dataset, which describes the transcripts expressions in SCLC, LUSC, and LUAD samples, was obtained from GEO and were analyzed by GEO2R tool, Venn diagram and Metascape.10,11
Statistical analysis
The differences between groups were determined by using The Statistical Package for the Social Sciences, version 26 (IBMCorp, Armonk, NY, USA). Receiver operating characteristic (ROC) curves were used to evaluate the performance of diagnostic tests. Kaplan-Meier was used to create the overall survival (OS) curve.
Results
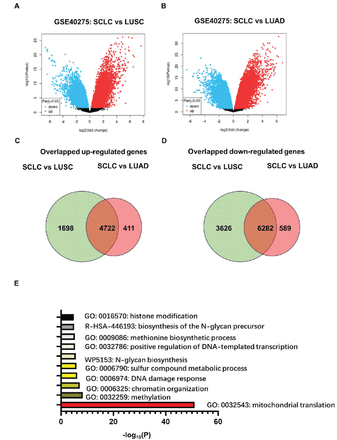
The GSE40275 dataset mapped the global transcriptome sequencing results of tissue samples derived from SCLC and NSCLC (mainly including LUSC and LUAD).10 With an adj. p-value of <0.05, we identified 16323 differentially expressed targets between the SCLC and LUSC (6420 up-regulated and 9908 down-regulated), and 12004 differentially expressed targets between the SCLC and LUSC (5133 up-regulated and 6871 down-regulated; Figure 1A&B). We found that there were only 330 differentially expressed genes between LUSC and LUAD. Above results indicated significant genetic differences both between SCLC and LUSC and between SCLC and LUAD, while molecular phenotypes of LUAD and LUSC are relatively similar.
- Differential transcripts between small cell lung cancer (SCLC) and lung squamous cell carcinoma (LUSC), and between SCLC and lung adenocarcinoma (LUAD), and the overlapping targets based on the GSE40275 dataset. The volcano maps showing the differential transcripts between A) SCLC and LUSC; and B) between SCLC and LUSC, based on the GSE40275 dataset. The GEO2R tool was used to identify differentially expressed genes. The Venn diagram showing the overlapped C) up-regulated transcripts; and D) down-regulated transcripts among the differential transcripts described in A & B. E) GO enrichment of the overlapped differentially expressed transcripts described in C & D. The plot was calculated and drawn by Metascape Gene List Analysis. LUSC: lung squamous cell carcinoma, LUAD: lung adenocarcinoma, SCLC: small cell lung cancer
The Venn diagram showed 11004 overlapping targets between the 2 comparisons (4722 up-regulated and 6282 down-regulated; Figure 1C&D). The main biological processes related to the overlapping targets were mapped using GO analysis (Figure 1C), and it indicated that these targets were highly involved in the mitochondrial translation, DNA or RNA methylation and methionine biosynthetic process (Figure 1E). Therefore, we focused on the genes that were involved in both mitochondrial translation and DNA or RNA methylation.
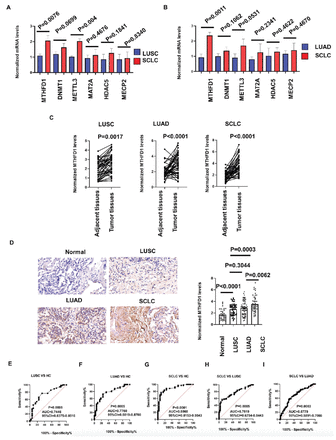
We next used a small-scale samples to quickly verify the bioinformatics analysis results above. We included 18 paired tissues (tumor tissues and matched adjacent tissues) from 6 cases of SCLC, 6 cases of LUSC and 6 cases of LUAD, who were randomly selected from participants in cohort 1. Total RNAs were extracted from all the tissues above, and extract their total RNAs. We focused on DNMT1, MAT2A, HDAC5, METTL3, MECP2, and MTHFD1 because they are all involved in both mitochondrial translation and DNA or RNA methylation, and ranked high in differential expression analysis. We found that MTHFD1, DNMT1, and METTL3 were higher in SCLC tissues than those in LUSC and LUAD tissues, and MTHFD1 has the greatest expression difference (Figure 2A&B). Therefore, we next explore the role of MTHFD1 using a large sample size.
- MTHFD1 was higher expressed in small cell lung cancer (SCLC) tissues than lung squamous cell carcinoma (LUSC) and lung adenocarcinoma (LUAD) tissues. Comparing the mRNA levels of DNMT1, MAT2A, HDAC5, METTL3, MECP2, and MTHFD1 between: A) SCLC tissues (n=6) and LUSC tissues (n=6); and B) between SCLC tissues (n=6) and LUAD tissues (n=6) in a small-scale samples. C) Comparing the MTHFD1 levels between tumor tissues and matched adjacent tissues in LUSC (left, n=56), LUAD (middle, n=70), and SCLC (right, n=70) cases in a large-scale samples. D) Comparing the MTHFD1 levels in tissues from healthy controls (HCs, n=33), LUSC (n=56), LUAD (n=70), and SCLC (n=70) cases in a large-scale samples. E-I) The receiver operating characteristic curve for the MTHFD1 levels in distinguishing: E) LUSCs and HCs; F) LUADs and HCs; G) SCLCs and HCs; H) SCLC and LUSCs; and I) SCLC and LUADs. Left: representative stained images among these 4 groups, right: bar chart comparison of differences among these 4 groups, HCs: healthy controls, LUSC: lung squamous cell carcinoma, LUAD: lung adenocarcinoma, SCLC: small cell lung cancer
We included 196 LC patients in cohort 1 to delve into the expression characteristics of MTHFD1. Detailed demographics for all the subjects are listed in Table 1. We also enrolled 33 gender and age matched HCs. A total of 56 patients were LUSC, 70 were LUAD, and 70 patients were SCLC.
- Clinical indications for included lung cancer patients.
Immunohistochemical results show that the MTHFD1 increased in cancer tissues compared with the adjacent tissues, regardless of their cancer tissue types, including SCLC, LUSC, and LUAD (Figure 2C). Meanwhile, we confirmed that the expression of MTHFD1 in all the LC types were significantly higher than that in HCs, and it was particularly upregulated in SCLC, compared to LUSC and LUAD (Figure 2D).
Receiver operating characteristic curves were next constructed. The MTHFD1 levels discriminated LUSC, LUAD, and SCLC patients from healthy control patients (Figure 2E-G). And, MTHFD1 could discriminate SCLC from LUSC and LUAD patients (Figure 2H&I). These results indicated that MTHFD1 might act as a biomarker for the diagnosis of both NSCLC and SCLC, and could discriminate SCLC from NSCLC.
Next, SCLC, LUSC, and LUAD patients were allocated into MTHFD1 high group and low group. Among 70 SCLC, 56 LUSC and 70 LUAD patients, 31 SCLC patients, 22 LUSC patients and 25 LUAD patients were MTHFD1 high phenotype and others were MTHFD1 low cases. For patients with SCLC, MTHFD1 high phenotype was associated with T1 stage and distant metastasis. However, MTHFD1 high phenotype was not associated with any indicators in LUSC and LUAD patients (Table 2).
- Association of MTHFD1 levels with clinical indications for small cell lung cancer, lung squamous cell carcinoma and lung adenocarcinoma patients.
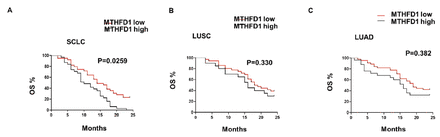
Next, we explored the relationship between MTHFD1 levels and LC prognosis after CRT treatment. We used cohort 2, which were all from cohort 1, except for excluding the non-CRT treated in cohort 1. The results showed a visible worse OS rate in the SCLC group, compared with both LUSC group and LUAD group. And, for the patients with SCLC, MTHFD1 high phenotype was associated with a poor 2-year OS curve (Figure 3A), whereas for the patients with LUSC and LUAD, the 2-year OS curve was not significantly associated with the MTHFD1 levels (Figure 3B&C). Collectively, these results indicated that MTHFD1 levels were specifically associated with prognosis of SCLC.
- MTHFD1 levels were specifically associated with prognosis of small cell lung cancer (SCLC) patients after chemoradiotherapy treatment. Kaplan-Meier curves for time to 2-year overall survival of patients with: A) SCLC; B) lung squamous cell carcinoma; and C) lung adenocarcinoma according to MTHFD1 levels. LUSC: lung squamous cell carcinoma, LUAD: lung adenocarcinoma, SCLC: small cell lung cancer, OS: overall survival
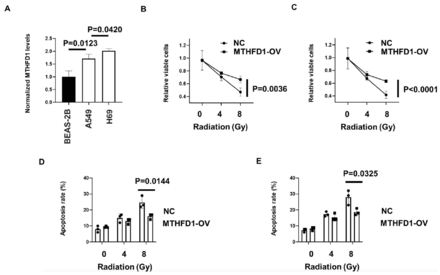
The MTHFD1 expression was subsequently analyzed by ELISA in cultured cell lines of SCLC (H69), NSCLC (A549) and normal lung epithelial cells (BEAS-2B). We found that it was increased in cancer cell line, and were expressed higher in H69 than A549, which is in accordance with clinical findings (Figure 4A). To determine whether MTHFD1 regulated the radio-sensitivity of SCLC cells, the viability and cell apoptosis of H69 and A549 after exposure to various doses of irradiation was analyzed. Cell viability was inhibited (Figure 4B&C), and cell apoptosis was induced (Figure 4D&E), by irradiation in a dose-dependent manner, and overexpression of MTHFD1 led to a markedly lower level of radiation-induced cell death and cell apoptosis, compared with the control cells, in both H69 and A549 cells (Figure 4B-E). These results indicated that MTHFD1 increases radio-resistance in both SCLC and NSCLC cells.
- Overexpression of MTHFD1 increases radio-resistance in both small cell lung cancer and non-small cell lung cancer in vitro. A) MTHFD1 levels were determined in H69, A549, and BEAS-2B cells. B-C) The H69 and A549 cells were instantly transfected with MTHFD1 over-expressed plasmid for 24 hours, and then the viable cells of B) H69; and C) A549 were detected after treated by different dose irradiation. D-E) The H69 and A549 cells were instantly transfected with MTHFD1 over-expressed plasmid and control plasmid for 24 hours, and then the apoptosis rate of B) H69; and C) A549 were detected after treated by different dose irradiation.
Discussion
Biomarkers are needed that enable earlier detection and monitoring of LC.12 The RNA-seq data for analyzing LC samples can accelerate the development process of biomarker. For example, based on the integrating single-cell RNA-seq and bulk RNA-seq, Zhang et al13 identified new therapeutic targets for patients with advanced LUAD. Similarly, Lavanya et al14 screened a series of potential specific biomarkers for SCLC by mining existing RNA-seq data. The GSE40275 dataset provides RNA-seq data for different tissue subtypes such as SCLC, LUSC, and LUAD, and some groups have made progress in LC research by analyzing this dataset.10,15,16 The starting point of this study is to screen targets by comparing the RNA-seq data between SCLC and LUSC, as well as between SCLC and LUSC, and taking their intersections, which will increase the probability of discovering SCLC specific biomarkers. After that, we screened many targets highly involved in mitochondrial translation, DNA or RNA methylation and methionine biosynthetic process.
The MTHFD1 ranked high among the targets, and was confirmed by us to be up-regulated in SCLC, which could discriminate SCLC from LUSC and LUAD. The MTHFD1 was the primary source of folate-activated one-carbon units in the cytoplasm. Increased evidence has been prompted that MTHFD1 was responsible for variable tumor. Yu H et al7 confirmed that overexpression of MTHFD1 in hepatocellular carcinoma could predict poorer survival and recurrence. In our study, we found that high MTHFD1 levels were specifically associated with poor prognosis of SCLC, but not with LUAD and LUSC, after CRT treatment. In terms of mechanism, MTHFD1/folate/one-carbon network is required for the de novo synthesis of 3 of the 4 DNA bases and the re-methylation of homocysteine to methionine, and therefore exerts its effects in tumor cells.17,18 It is reported that aberrant DNA methylation, causing gene silencing, is common in cancers and would induce CRT resistance.19 And, some studies have reported the significant differences in methylation levels between SCLC and NSCLC.20 Correspondingly, our clinical data and experimental data showed that the expressions of MTHFD1 in SCLC cells are significantly higher than that in NSCLC cells, and high expression of MTHFD1 reduces the sensitivity of radiation to cancer cells.21 These might be the major reason why MTHFD1 levels can discriminate SCLC from LUSC and LUAD, and specifically reflect the prognosis of SCLC.
Study limitations
this work is just an initial exploratory study, so it has limitations due to a relatively small sample size. We are carrying out a broader study with a more diverse range of samples provided by multiple medical centers to substantiate our findings.
In conclusion, high MTHFD1 expressions could distinguish SCLC from LUSC and LUAD, and was associated with poor CRT treatment outcome of SCLC. The implication of this study is that it provides a new understanding of the relationship between MTHFD1 expression and LC progressions, indicating that MTHFD1 levels might be a specific indicator of SCLC.
Acknowledgment
The authors gratefully acknowledge Editage (https://www.editage.cn/) for their English language editing.
Footnotes
Disclosure. Authors have no conflict of interests, and the work was not supported or funded by any drug company.
- Received June 22, 2024.
- Accepted July 4, 2024.
- Copyright: © Saudi Medical Journal
This is an Open Access journal and articles published are distributed under the terms of the Creative Commons Attribution-NonCommercial License (CC BY-NC). Readers may copy, distribute, and display the work for non-commercial purposes with the proper citation of the original work.