Abstract
Objectives: To identify the carbapenemase producing Gram-negative bacteria (GNB) by phenotypic methods and to confirm the presence of resistant genes using real-time polymerase chain reaction (PCR).
Methods: This was a prospective study carried out at the Department of Microbiology, Sri Venkata Sai Medical College and Hospital, Mahabubnagar, India, from March 2018-2021. All samples were screened for carbapenem resistance by disc diffusion method and the VITEK®2 compact system (bioMérieux, France). Detection of carbapenemase was carried out using RAPIDEC®CARBA NP test (Biomeriux Private Limited, South Delhi, India), screening for metallo-β-lactamases (MBL) was carried out by double disk synergy test (DDST), and genotypic characterization by real-time PCR.
Results: Among the 1093 Gram-negative bacilli identified, 220 (17.0%) were resistant to carbapenems by both tested methods. Carbapenemase detection using the RAPIDEC®CARBA NP test indicated that 207 (94.0%) were carbapenemase producers, of which 189 (91.2%) were MBL producers. The most common carbapenemase genes identified were New Delhi metallo-β-lactamase (NDM; 47.3%), followed by the co-existence of genes in combination of NDM, with Verona integron-mediated metallo-β-lactamase (VIM; 39.6%), VIM and oxacillin hydrolyzing enzymes-48 (OXA-48; 4.3%), and OXA-48 (1.4%).No gene of active on imipenem, Klebsiella pneumonia carbapenemase, VIM, or OXA-48 alone was detected.
Conclusion: This study suggests routine carbapenem resistance testing among multi-drug resistant-GNBs, as most of these infections occur in hospitals. In addition, there is a possibility that these highly antibiotic-resistant genes could spread to other bacteria resulting in further dissemination.
Hospital-acquired infections (HAI) are defined as infections that occur after 48 hours or more of hospital entry. Hospital infections include; central line-associated bloodstream infections, ventilator-associated pneumonia, catheter-associated urinary tract infection, and surgical site infection. The major contributors to hospital-acquired infections include several factors such as, prolonged hospital stay, invasive measures, ventilator support, immune suppression, old age, diabetes, and stay in the intensive care unit, indwelling devices, and recurrent visits to hospitals. Morbidity and mortality are the most important challenges associated with HAI.1,2
Carbapenem drugs are the most valuable drugs for treating multi-drug resistant Gram-negative bacteria (MDR-GNB) infections and have been used for the past 10 years. However, there has been a significant growth of carbapenem-resistant organisms that cause severe damage to public health.3,4 These carbapenems, which include meropenem, ertapenem, and imipenem, are β-lactam antibiotics that possess a β-lactam ring and have a wide range of activity and great usefulness. These antibiotics are used as a last line of defense for treating infections caused by MDR-GNBs as well as organisms that produce extended-spectrum β-lactamases, which are used to screen carbapenem resistance in laboratories, and develop resistance either through gene transfer or mutations.5,6
Carbapenem resistance is mainly owing to the expression of a carbapenemase enzyme, efflux pump, or porin loss. Among these, the most important and difficult mechanism is the production of the carbapenemase enzyme, because it is present on mobile genetic elements, which are easily transferable from one bacterium to another bacterium such as Pseudomonas spp., Acinetobacter spp., Escherichia coli (E. coli), and Klebsiella spp., which the World Health Organization (WHO) has designated as high priority organisms in 2017.7-10 The major carbapenemase genes are bla-Klebsiella pneumonia carbapenemase (blaKPC), bla-oxacillin hydrolyzing enzymes-48 (blaOXA-48), bla-New Delhi metallo-β-lactamase (blaNDM), bla-Verona integron-mediated metallo-β-lactamase (blaVIM), and bla-active on imipenem (blaIMP), which are present globally and cause nosocomial infections. Many researchers have studied various methods of carbapenem resistance detection including carbapenemases.8,11,12 Methodologies for the detection of carbapnem resistance range from conventional disc diffusion and minimum inhibitory concentration (MIC) determination methodologies to advanced and rapid detection methods such as automated methods, RAPIDEC®CARBA NP (Biomeriux Private Limited, South Delhi, India), and genotypic methods using polymerase chain reaction (PCR). Many of these advanced methodologies are not routinely used in peripheral and resource poor settings because of which detection of these resistant organisms is missed or delayed which in turn, leads to therapeutic failure and dissemination of these resistant strains in the hospitals. Identifying the prevalence of these carbapenem-resistant Gram-negative bacilli (CRGNB) in any hospital will help to formulate the appropriate infection control guidelines which prevent further spread of these infections and also economical burden on the hospitals.
As a result, we used genotypic approaches to estimate the prevalence of β-lactamase Gram-negative microorganisms CRGNB and to identify carbapenem-producing isolates, which are important in choosing empirical therapy, designing good antibiotic policies, updating local antibiotic guidelines for doctors, and in determining clinical treatment failure. With this background, this study aimed to screen for carbapenemase production among HAI using both phenotypic and genotypic methods.
Methods
This was a prospective study carried out in the Department of Microbiology, Sri Venkata Sai (SVS) Medical College and Hospital, Mahabubnagar, India, over a period of 3 years from March 2018-2021. A certification of Ethical Clearance (SVS Medical College/Institutional Ethical Committee; approval no.: 05/2018-623) was obtained and the study was carried out in accordance to the principles of Helsinki Declaration.
All the inpatient’s samples with suspected HAI showing growth of laboratory confirmed CRGN bacteria such as E. coli, Klebsiella spp., Acinetobacter spp., Pseudomonas spp., and Enterobacter spp. isolates were included in the study. Samples with carbapenem sensitive organisms, clinically insignificant growth characters, and other organisms other then the mentioned were excluded from the study.
A total of 1093 Gram-negative bacteria that were isolated from various clinical samples, such as urine, pus, stool, sputum, blood, endotracheal secretions, cerebrospinal fluid, and other body fluids from hospitalized patients were processed at the Department of Microbiology, SVS Medical College and Hospital and were identified to the species level based on the Gram staining, colony morphology, and biochemical reaction. These isolates were screened for carbapenem resistance using the Kirby Bauer disc diffusion method.
The results were interpreted according to the Clinical Laboratory Standards Institute (CLSI-M1OO-S29) guidelines.13 Organisms that were resistant to imipenem and meropenem were further processed to determine their MICs using the VITEK®2 compact system (bioMérieux, France).14 Antibiotic resistance was detected for antibiotics such as cefoxitin, cefepime, ceftazidime, ceftriaxone, piperacillin, tazobactam, imipenem, meropenem, ciprofloxacin, ertapenem, gentamicin, amikacin tigecycline, colistin, cefoperazone sulbactum, and cotrimoxazole (Himedia, Mumbai, India). Klebsiella pneumonia ATCC BAA 1705 and ATCC BAA 1706, Escherichia coli ATCC 25922 were used as the positive and negative controls for phenotypic and genotypic investigations.
Detection of carbapenems production
Imipenem and meropenem resistant organisms were used in further investigations.
RAPIDEC® CARBA NP test
The test was carried out as described by Nordmann, Poirel, and Dordet “A change in color of the Ph indicator is indicative of carbapenem hydrolysis caused by carbapenemase-producing bacteria, which produce acid”.15 The procedure followed the manufacturer’s instructions. API suspension medium (25 μL; bioMérieux, New Delhi, India) was added to the wells and 5-6 colonies from the fresh culture plate were obtained and added to the prescribed well. The turbidity of the inoculum was compared with the provided strip. Next, 10 μL of the inoculum was added to 2 wells of which one well contained imipenem. Imipenem was used as the carbapenemase zinc substrate for metallo-β-lactamases (MBL)-producing gram-negative bacteria and the results were considered positive if the color changes from red to yellow, orange, or thick orange in comparison to the control well. Organisms that tested positive for carbapenemase were used for further evaluations.15
Double disk synergy test
Phenotypic testing was carried out following the guidelines of the CLSI. Briefly, 0.5 Mac Farland bacterial inoculum was spread on a Muller Hinton Agar plate as a lawn culture and one imipenem disk was placed at the center with a blank disc place adjust 20 mm apart. Next, 10 μL of 0.5 ethylene-diamine-tetraacetic acid (750μg) was inoculated on the blank disc and incubated at 37°C for 24 hours. Presence of an inhibitory zone was considered MBL-positive, and cultures that did not exhibit such zones were considered as the Serine group.16
Genotyping method
The overnight bacterial culture was used for DNA extraction using the Hi Per Bacterial Genomic DNA Extraction Kit (Hi-Media HTBM009) in accordance with the manufacturer’s instructions.
Bacteria DNA was isolated and eluted from the columns in 200 μL elution buffer and stored in a mini Eppendorf tube at -20°C until further use.
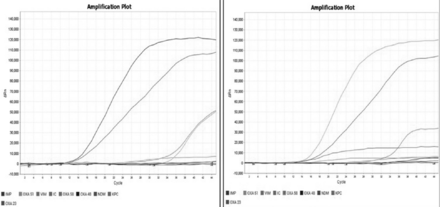
The carbapenemase gene (multiplex) probe-based Hi-Media Hi-PCR kit was used to detect specific regions of the gene encoding the carbapenemase enzyme. The technique is very easy, fast, and powerful for the detection of carbapenemase genes such as blaVIM, blaNDM, blaKPC, blaOXA-48, and blaIMP and can be used to accurately identify one or a combination of carbapenemase genes in a single tube reaction with a large variety of organisms. Positive, internal, and negative controls were used as specified in the manufacturer’s instructions. Briefly, the multiplex probe PCR kit is designed to detect specific regions of the genes encoding various carbapenemase enzymes. There are 2 master mixes in this kit, wherein master mix-1 detects NDM, KPC, IMP, and VIM in the FAM, HEX, Texas Red, and Cy5 channels, where as master mix-2 detects OXA-51, OXA-23, OXA-48, and OXA-58 in the FAM, HEX, Texas Red, and Cy5 channels. The internal control was detected in the Cy5.5 channel in both the master mixes. The kit allows sensitive and specific detection of single and co-present carbapenemase-encoding genes in a single tube reaction. The cycling method consisted of initial denaturation at 95°C for 10 minutes, followed by denaturation at 95°C for 5 seconds, and 45 cycles of annealing and extension at 60°C for one minute, and the final holding stage was carried out in Quant Studio real-time PCR (ThermoScientific). A cycle threshold value of ≤40 and band was considered positive (Figure 1).
- Representing amplification plot of New Delhi metallo-β-lactamase (NDM), Verona integron-mediated metallo-β-lactamase (VIM), and oxacillin hydrolyzing enzymes-48 (OXA-48) by using Hi-media HiPCR carbapenemase gene (multiplex) probe PCR kit; identification of genes using real-time polymerase chain reaction.
Statistical analysis
Data obtained were analyzed using descriptive statistics, Chi-square test carried out using Statistical Packaage for the Social Sciences, version 23.0 (IBM Corp., Armonk, NY, USA). A p-value of <0.05 was considered significant.
Results
Among the 1093 Gram-negative bacilli identified, 220 (17%) by using the disc diffusion method and VITEK®2 methods, they were shown to be resistant to carbapenems. The carbapenemase enzyme was detected in these species utilizing the RAPIDEC®CARBA NP test, in which 207 (94.0%)were identified as carbapenemase producers. Among these, 189 (91.2%) were MBL producers and 18 (8.6%) were non-MBL producers by the double disk synergy test (DDST) method.
Among all the carbapenemase producers, the pre-dominant organisms were Klebsiella spp. (56.7%), followed by E. coli (17.7%), Acinetobacter spp. (11.4%), Pseudomonas spp. (10.9%), and Enterobacter (3.1%).
In the 207 carbapenemase-producing isolates, carbapenemase genes were detected in 192 (92.7%) isolates using real-time PCR. The most common genes identified were NDM (47.3%), followed by the co-existence of genes in combination of NDM, with VIM (39.6%), VIM and OXA-48 (4.3%), and OXA-48 (1.5%). No gene of IMP, KPC, VIM, or OXA- 48 alone was detected.
Among the NDM gene-positive organisms, Klebsiella (58.2%) was the most common organism, followed by E. coli (22.4%), Acinetobacter (9.2%), Pseudomonas (8.2%), and Enterobacter (2.0%). The co-existence of VIM and NDM appeared predominantly in Klebsiella (57.3%), followed by E. coli (14.6%), Acinetobacter (12.2%), Pseudomonas (11.0%), and Enterobacter (4.9%). Klebsiella and Pseudomonas exhibited the co-existence of OXA-48 and NDM, and 3 genes (NDM, VIM, and OXA-48) were co-expressed in Klebsiella (44.4%), Acinetobacter (33.3%), and Pseudomonas (22.2%) isolates (Table 1).
- Prevalence of genes in different carbapenem-resistant isolates.
The majority of the organisms were isolated from endotracheal secretions (23.9%), followed by pus and wound swabs (21.3%), blood (22.3%), urine (8.8%), sputum (13.0%), stool (7.8%), cerebrospinal fluid (1.5%), and other body fluids (1.0%). The most predominant organisms in endotracheal secretions were Klebsiella (58.7%) and Acinetobacter (15.2%). The predominant organisms in the pus samples were Klebsiella (65.9%), followed by Pseudomonas (17.1%). In urine samples, E. coli was predominant (41.2%), whereas, in sputum samples it was Klebsiella (80.0%) and in stool samples, it was E. coli (80.0%). Based on statistical analysis, it was found that the prevalence of carbapenems-resistant isolates differed significantly across other type of samples (p<0.05; Table 2).
- Distribution of carbapenemase gene-producing organisms in different samples.
The prevalence of carbapenemase genes was higher in the 0-21 years age group with 88 (45.8%) cases followed by the 21-40 years with 52 (27%) cases.
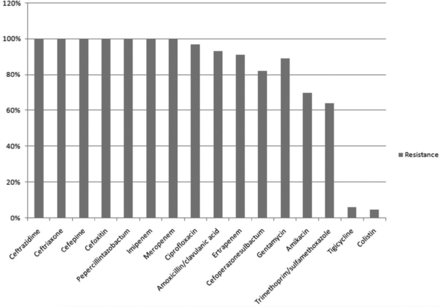
There was 100% resistance to cefoxitin, cefepime, ceftazidime, ceftriaxone, piperacillin-tazobactam, imipenem, and meropenem. Resistance to further antibiotics was varied, ciprofloxacin (96.7%), ertapenem (91.0%), gentamicin (89.0%), amikacin (69.6%), cefoperazonesulbactam (82.0%), trimethoprim/sulfamethoxazole (64.0%), tigecycline (6.14%), and colistin (4.7%). A important variance was detected in the resistance of CRGN bacteria to numerous antimicrobial agents (p<0.05; Figure 2).
- Resistance pattern of isolates harboring carbapenemase genes. represents the resistance pattern to various drugs by carbapenemase producing Gram negative bacteria.
The pre-dominant genes in the blood isolates were NDM (55.8%), NDM and VIM (41.9%), followed by NDM, VIM, and OXA-48 (2.3%). The dominant gene in cerebrospinal fluid samples was NDM and VIM (66.7%), followed by NDM (33.3%). In endotracheal secretions, the co-existence of NDM and VIM was prevalent (45.7%), followed by that of NDM (41.3%).In pus and wound swabs, 19 (46.3%) of the most prevalent genes were NDM and NDM with VIM. In sputum, the most common gene isolated was NDM (52.0%), followed by NDM and VIM (44.0%). In stool, the pre-dominant gene was NDM (80.0%; Table 3).
- Distribution of carbapenemase genes in different samples.
Discussion
Carbapenem is the medication of choice for the treatment of extended-spectrum β-lactamases and MDR organisms. Resistance to carbapenems is progressively being observed, especially in HAI, which are difficult to treat, pose a huge economic burden and are allied with improved mortality and morbidity.17
To decrease the threat of infection spreading in the hospital and to reduce mortality rates, this study re-connoitered the existence of carbapenemase-producing organisms using phenotypic methods and determined the distribution of carbapenemase genes by genotypic testing. The prevalence of carbapenem resistance was 17.0% in this study, which is similar to Haji et al18’s study. The incidence of prevalence in various parts of India varies from 14-69%. This was mostly owing to infection control practices, hospital infrastructure, and the number of antibiotics used.19
One of the major goals of this research was to see whether carbapenemase-genes were present in gram-negative bacteria isolated form hospitalized patients. Carbapenemase-producing micro-organisms are difficult to detect and require phenotypic and genotypic analyses. Of the 220 isolates, 207 (94.0%) had phenotypic positivity, which was comparable to that reported by Diwakar et al.20 Our genotypically positive findings (87.2%) were comparable to the genotypically positive rate of 90.3% reported by Garg et al.21 The major differences were likely due to geographic region, the testing method used, and the organism.22
New Delhi metallo-β-lactamase (47.3%) was the pre-dominant gene in this study, which was similar to that reported by Naim et al.23 The prevalence of resistance genes was higher in Klebsiella spp. (56.7%) followed by E. coli (17.7%). The prevalence of NDM in India is causing major public health challenges due to its elevated medical and economic burden.24 We found that blaKPC, blaSIM, and blaIMP genes were not detected, which is consistent with the observations made by Garg et al.21
In this study, the co-existence of VIM and NDM was 39.6%, whereas Mohanam et al25 reported 14.6% and Ellappan et al26 reported a 17.3% co-existence in Pseudomonas aeruginosa from Southern India.
The co-existence of NDM and OXA-48 in this study was 1.4%, whereas in a study by Garg et al21 it was 20.0%. Grag et al21 identified the NDM and OXA-48 co-existance pre-dominantly in E.coli, followed by Klebsiella, and Enterobacter. None of the nonfermenters in his study exhibited OXA-48 gene. Whereas, according to Vatansever et al27 the co-harboring of OXA-48 and NDM in colistin-resistant Pseudomonas aeruginosa was 88.8%. Males were more likely to be resistant to carbapenem (66.8%), similar to that reported by Esther et al.28 New Delhi metallo-β-lactamase prevalence in stool samples was approximately 7.8%, which was slightly higher than the 3.6% reported by Pan et al29 and lower than the 18.5% reported by Esther et al.28 Moreover, these results confirmed that stool samples were colonized with carbapenemase genes, thus emphasized the need to screen for NDM.28,29 A large proportion of carbapenem-resistant organism’s exhibit resistance to commonly used drugs. Most CRGNBs are still sensitive to tigecycline and colistin, which are the last lines of defense against CRGNB. We found that 18 (8.6%) of the samples we evaluated were phenotypically positive. However, no genes were detected using real-time PCR. There may be a combination of factors contributing to this effect, such as gene targets in this study, a mutation, or loss of porin. Thus, it is important to accurately identify the genes responsible for carbapenemase production for better treatment outcome and also further research needs to be carried out to identify all the mechanisms of resistance co-existing in the resistant isolates, which if identified correctly can help in better management of patients by choosing appropriate antibiotics either alone or in combination for the treatment of these drug resistance infections.
Study limitation
The study was a single center study carried out in a tertiary care center and only resistance to carbapenems by carbapenmase production was studied. Role of other co-existing mechanism such as porin loss or efflux pumps in the causation of carbapenem resistance has not been evaluated.
In conclusion, carbapenemase genes are spreading rapidly worldwide due to the increased prevalence of horizontal transfer. As there are no new drugs available, and prevalence incidences vary regionally, real-time PCR probe-based detection of these genes is beneficial for early detection, developing infection control protocols, and promoting appropriate antibiotic use. The presence of co-existing carbapenemase genes is concerning. In addition, there is a possibility that highly antibiotic-resistant genes could spread to other bacteria because of their high spreading ability and thereby increase the possibility of further dissemination. Routine carbapenem resistance testing is suggested among MDR-GNBs at least in HAI as the prevalence was high which was shown in this study.
Acknowledgment
The authors gratefully acknowledge Manipal Academy of Higher Education, Karnataka, Palamur Biosciences Pvt. Ltd., and SVS Medical College, Mahabubnagar, India, for thier cooperation. We also would like to thank Dr. Murali for providing his comments on the draft version of the manuscript and Dr.Sreekanth for helping in the manuscript writing. We would like to thank Editage for English language editing.
Footnotes
Disclosure.This study was funded by the Department of Science and Technology, Government of India, New Delhi, India, as part of the Women Scientist Scheme (no.: SR/WOS-A/LS-643/2016 [G]).
- Received October 8, 2021.
- Accepted February 15, 2022.
- Copyright: © Saudi Medical Journal
This is an Open Access journal and articles published are distributed under the terms of the Creative Commons Attribution-NonCommercial License (CC BY-NC). Readers may copy, distribute, and display the work for non-commercial purposes with the proper citation of the original work.