Abstract
Objectives: To assess the oxidation state and gene expression profiles of relevant enzymes in intrauterine growth restriction (IUGR) patients in Saudi Arabia.
Methods: Current case-control study involved plasma and placental tissue samples from 25 IUGR patients and 25 healthy pregnant (HP) women attending the Obstetrics and Gynecology Clinic, King Khalid University Hospital, Riyadh, Saudi Arabia, between April and November 2017. We compared hydrogen peroxide, superoxide anions, malondialdehyde, and oxidative stress markers levels and the activities of glutathione-related enzymes (glutathione peroxidase [GPx], glutathione reductase [GR], glutathione S-transferase [GST], glutamate cysteine ligase [GCL], glutathione synthetase [GS], reduced glutathione [GSH], oxidized glutathione [GSSG], and oxidized nicotinamide adenine dinucleotide [NAD+], and reduced NAD [NADH]) between the 2 groups. We also compared differential expression levels of glutathione-related enzyme genes using reverse transcription-quantitative polymerase chain reaction.
Results: Oxidative stress markers significantly differed in IUGR samples, while GSH levels and GPx, GR, GST, GCL, and GS activities and their placental mRNA transcriptional levels were significantly lower. Plasma and placental NAD+ levels were also significantly lower, while NADH levels were significantly higher, causing lowered NAD+-NADH ratios in the IUGR group compared to control.
Conclusions: Intrauterine growth restriction patients show a metabolic shift in favor of oxidation compared to HP women.
Intrauterine growth restriction (IUGR) is a common complication affecting 10%-15% of pregnancies worldwide and is also known as small gestational age.1 Intrauterine growth restriction is a condition by which the fetal weight is estimated at or below the 10th percentile for the corresponding pregnancy stage. It is a significant causative factor for perinatal complications and a significant public health issue.2 In addition, IUGR is also a major risk factor for fetal death and accounts for 29.3% of all fetal death cases in Saudi Arabia.3
Intrauterine growth restriction of placental origin occurs early in pregnancy (during trophoblast invasion into the decidua). To support the cell proliferation, growth, and metabolism, high mitochondrial activity and biogenesis are required to regulate placental metabolic activity, because mitochondria are the main source of cellular energy.4 Therefore, any mitochondrial defect may lead to increased reactive oxygen species (ROS) generation, some of which are highly toxic and cause placental damage.5
Physiologically, ROS generated by various cellular mechanisms function as secondary messengers and holding crucial role in regulating several cellular mechanisms.6 However, ROS levels exceeding the limits that a cell can manage by releasing antioxidants can significantly damage cell organelles or cause major structural defects in critical cell components, such as lipids of the cell membrane, proteins, and DNA.5 An imbalance in normal cell homeostasis can result in oxidative stress (OS). Therefore, maintaining a balance between the ROS generated and the released antioxidant levels is crucial for normal cell homeostasis.
In pregnancy, an imbalance in normal redox status leads to OS, which is crucial in dysfunction of the placenta such as IUGR in which the maternal blood supply to the placenta is disturbed.7 Although the mechanism underlying IUGR is unclear, OS might be a potential cause. However, the association between OS and IUGR pathogenesis, especially in Saudi Arabia, has not been fully described. In addition, the gene expression of glutathione-related enzymes relative to the ROS status, especially in IUGR patients, has not been reported. Therefore, this study determined the plasma and placental levels of OS markers/lipid peroxidation (hydrogen peroxide [H2O2], superoxide anions [SOA], malondialdehyde [MDA]) and oxidized glutathione (GSSG) in IUGR patients and healthy pregnant (HP) women. We also measured the activities of glutathione-related enzymes, including glutathione peroxidase (GPx), glutathione reductase (GR), glutathione-S-transferase (GST), glutamate cysteine ligase (GCL), and glutathione synthetase (GS), and the plasma and placental levels of reduced glutathione (GSH), nicotinamide adenine dinucleotide (NAD+), and reduced NAD (NADH) in IUGR patients and gestational-age-matched HP women. In addition, we profiled differential expression placental levels of glutathione-related enzyme genes in IUGR patients and gestational-age-matched HP women.
Methods
The present case-control study enrolled 25 HP Saudi women as controls and 25 IUGR patients. Samples were collected from the Obstetrics and Gynecology Clinic, King Khalid University Hospital, Riyadh, Saudi Arabia between April and November 2017. All participants were primigravidae (pregnant for the first time) and had similar demographic backgrounds. The HP group was matched by body mass index (BMI), gestational age, and the age to the IUGR group.
Prior to sample collection, consent forms were signed by all participants. The study was permitted by the Research Ethics Committee of the College of Applied Medical Sciences, Riyadh, King Saud University (CAMS 093-37/38). The study was performed in accordance to the principles of Helsinki Declaration.
Inclusion and exclusion criteria
Healthy pregnant women with no history of pregnancy loss were treated as controls. Intrauterine growth restriction was defined as gestation-related birth weight under the 10th centile for the pregnancy stage. Diagnosis was determined by fetal ultrasound biometry and doppler ultrasound and confirmed on the basis of the IUGR infant’s birth weight compared to the controls (2.16 ± 0.23 kg versus 3.56 ± 0.34 kg; p<0.001) participants with diabetes mellitus, gestational diabetes, renal disease, hypertension, multiple pregnancies, chorioamnionitis, C-reactive protein ≥5mg/L, BMI ≥30 kg/m2, cardiovascular disease, hepatitis, and drug abuse were excluded from the study. In addition, participants with pregnancy diseases (preeclampsia, endometriosis, or polycystic ovary syndrome) were excluded because these diseases are associated with OS.
We collected 10mL of maternal venous blood samples in cold vacutainer EDTA tubes, and plasma obtained by 20 minutes centrifugation at 3000 × g then samples were stored at -80°C.
Trophoblastic tissue was excised from full-depth placental tissue, washed with phosphate-buffered saline (PBS; 0.1 M), and cut into ~1g fragments. The tissue pieces were placed in RNAlater® to stabilize RNA and then immediately snap-frozen in liquid nitrogen until gene expression profiling or in PBS for later biomedical assays. At the time of analysis, we homogenized the tissue in potassium chloride (0.1 M), then it underwent 10 minutes of 11000 × g centrifugation at 4°C, and the supernatant was used for assessment.
Oxidative stress markers assessment. Hydrogen peroxide generation rate
We measured H2O2 levels using 2’,7’-dichlorofluorescein diacetate (DCFH-DA), as reported previously.8 2’,7’-dichlorofluorescein diacetate is a non-polar molecule, and once inside cells, it is deacetylated to 2’,7’-dichlorofluorescein (DCFH) and oxidized to highly fluorescent substance of 2’,7’-dichlorofluorescein (DCF), under the action of H2O2. The fluorescence level of DCF was used as an indicator of the H2O2 generation rate.
Superoxide anions generation rate
We measured the SOA generation rate, as described by Al-Sheikh et al, 2019.8 Blue formazan was produced by reduced nitroblue tetrazolium (NBT), and its measurement was representative of SOA levels.
Lipid peroxidation and protein oxidation
Malondialdehyde levels resulting from lipid peroxidation were determined using thiobarbituric acid.5 We also determined protein oxidation levels by measuring the total protein carbonyl content (PCC), as previously described.9
Glutathione-related enzyme activity
We spectrophotometrically assayed plasma and placental GPx, GR, and GST activities using appropriate sample volumes, as previously described.5
We spectrophotometrically determined GCL and GS activities at 25°C by monitoring the adenosine diphosphate (ADP) formation rate by a compound assay with lactate dehydrogenase (LDH) and kinase pyruvate, as directed previously.10 The GCL 200 µL reaction solution was consistent of 100 mM buffer (Tris-HCL; pH 8.2), 5 mM adenosine triphosphate (ATP), 150 mMKCl, 20 mM MgCl2, 0.2 mM NADH, 10 mM L-cysteine, 2 mM EDTA, 2 mM phosphoenl pyruvate, kinase pyruvate (2U), and LDH (2U). The reaction solution of GS mixture consisted of 100 mM buffer (Tris-HCL; pH 8.2), 10 mM ATP, 50 mMKCl, 20 mM MgCl2, 0.2 mM NADH, 5 mM glycine, 5 mM L-α-glutamyl-L-α-aminobutyrate, 0.5 mM phosphoenl pyruvate, 2 mM EDTA, kinase pyruvate (5U), and LDH (8U). Reaction was initiated by mixing 50 µL of plasma and 75 µL of placental tissue supernatant to the reaction mix then monitored the absorbance rate decrease at 340nm. The specific activities of both enzymes were expressed in pmol/min/mL plasma for GCL and nmol/min/mL for GS, and in nmol/min/mg protein for placental GCL and GS.
Glutathione and GSSG assays
We measured GSH and GSSG levels in all samples by the recycling method of glutathione reductase-5, 5-dithiobis-2-nitrobenzoic acid (DTNB), as previously described.5
Nicotinamide coenzymes assay
We assayed plasma and placental NAD+ and NADH levels (30 µg protein), as described by Hyashi et al 2009.11 The pH of ice-cooled samples was balanced (6.5-7.5) and 0.5mL of glycylglycine (pH 6.5 or 7.5) was added to the coenzyme samples. Next, samples were placed into the reaction solution holding 50 mM glycylglycine, 0.5 mM EDTA (pH 7.4), 0.5 mM phenazine ethosulfate, 1 mM thiazolyl blue, and 65 µg/mL of alcohol dehydrogenase. Reaction was initiated mixing in ethanol (0.6M) and absorbance was recorded at 570 nm. Lastly, external and internal NAD+ levels were used to quantitate results, which were expressed in nM for plasma and µg/mL protein for placental tissue.
Gene expression profiling
Ribonucleic acid (RNA) extraction was performed followed by complementary DNA (cDNA) synthesis, and PCR, as described previously.12 We washed blood and the debris of placental samples with 0.9% saline and immediately transferred them into RNA later solution (Ambion, Austin, TX, USA). The samples were preserved at 4°C for 24 and later at -80°C for RNA analysis.
Gene expression using RT-qPCR
Preserved samples were thawed on ice and then processed for RNA total extraction and quantification of relative gene expression using reverse transcription-quantitative PCR (RT qPCR). Before analysis, we homogenized ~200 mg of trophoblastic and decidual placental samples separately for RNA isolation using TRIzol® (Invitrogen, Paisley, UK) and Tissue Lyser LT (Qiagen, Inc., Germany) and subsequently extracted total RNA according to the standard procedure. Next, we removed genomic DNA and synthesized cDNA using the SuperScript™ III First-Strand Synthesis for RT-qPCR SuperMix kit (Invitrogen). The total reaction volume was 25 µL containing 100 nM of the following gene primers; 5 µL of the cDNA sample, 12.5 µL of the SYBR Green PCR Master Mix S, 2.5 µL of the QuantiTect® Primer Assay (10X), and 2.5 µL of RNase free water: GCL (Hs_CAT_1_SG QuantiTect Primer Assay, QT00079674), GPx (Hs_GPx_1_SG QuantiTect Primer Assay, QT00203392), GR (Hs GSR_1_SG QuantiTect Primer Assay, QT00038325), GS (Hs_SOS_1_SG QuantiTect Primer Assay, QT01664327), and 18S (Hs_RRN18S_1_SG QuantiTect Primer Assay, QT00199367). We performed PCR and evaluated PCR amplicon specificity using 18S ribosomal RNA as an internal standard control, as previously described.13 We also performed TaqMan® gene expression assays (Applied Biosystems, Foster City, CA, USA) and determined the relative levels of gene expression (fold change [FC]) in relation to18S used as a housekeeping control gene by the 2-ΔΔCq method, as indicated previously.12
Statistical analysis
The statistical analysis was performed by the Statistical Package for Social Sciences, version 19.0 (IBM Corp, Armonk, NY, USA). Data normality was tested using Shapiro-Wilk test showing normal distribution (p>0.05). Hence, comparison of the levels of hydrogen peroxide, superoxide anions, malondialdehyde, OS markers levels, activities of glutathione-related enzymes, reduced glutathione, oxidized glutathione, oxidized nicotinamide adenine dinucleotide and reduced NAD (NADH), between the 2 groups was performed by t-test. Statistical significance was considered at p<0.05 and results were presented as the mean ± standard deviation (SD).
Results
The ages of the IUGR group (Table 1) range 20-29 years (26.2 ± 3.81) with a mean gestational age of 35.8 ± 2.98 weeks (range 33-37 weeks).
- Comparison of plasma and placental OS markers in IUGR and HP groups.
Plasma and placental oxidative stress marker levels
The mean plasma and placental levels of OS markers (H2O2, SOA, MDA, and PCC) found in the IUGR group were significantly higher in relation to the control (p<0.001 and p<0.0001; Table 1). The percentage increase in OS markers in the IUGR group was more pronounced, with 38.1%-50.3% magnitude in placental samples compared to 12.9%-15.1% in plasma samples.
Glutathione and GSSG levels and GSH-GSSG ratios
Reduced glutathione levels in the IUGR group were significantly lower (p<0.001). In addition, the decrease in placental GSH levels in the IUGR group was significantly more (p<0.0001) and was ~22% in placental samples compared to 12% in plasma. In contrast, plasma and placental GSSG levels were higher in the IUGR group in relation to the healthy control (p<0.001) and this was significantly higher in placental samples (p<0.0001); the increase was 16.9% in plasma and 30.7% in placental samples. The plasma and placental GSH-GSSG ratio significantly decreased in the IUGR (~40%) compared to the HP group (~35%; p<0.0001; Table 2).
- Plasma and placental GSH and GSSG levels and the GSH/GSSG ratio in IUGR and health pregnant (HP) groups.
Glutathione-related enzyme activity
Plasma and placental activities of GPx, GR, GST, GCL, and GS were significantly lower in the IUGR group and this decrease was more in plasma (p<0.001) than placental samples (p<0.0001; Table 3). The placental activity of all enzymes decreased with a similar magnitude (37%-52%) in the IUGR group relative to control. However, the activity of all enzymes decreased in plasma by only 12%-18% in the IUGR group relative to the healthy control.
- Glutathione-related plasma and placental enzyme activities in IUGR and healthy pregnant (HP) groups.
Nicotinamide coenzyme levels
Significant reduction (p<0.0001) was noted in the IUGR NAD+ levels, while NADH levels significantly increased relative to the HP group (p<0.0001). Therefore, plasma and placental NAD+-NADH ratio in the IUGR group was significantly low in relation to control (p<0.0001); the percentage decrease in the plasma ratio was 88% and placental NAD+-NADH ratio was 87% (Table 4).
- Plasma and placental nicotinamide coenzyme levels in IUGR and healthy pregnant (HP) groups.
Antioxidant enzyme gene expression
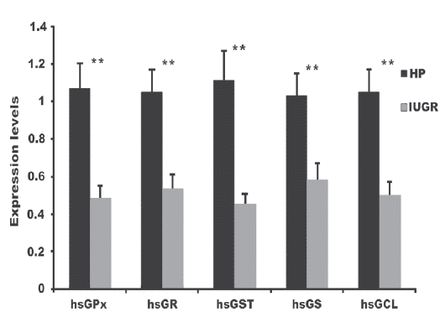
The messenger RNA (mRNA) expression significantly decreased for all investigated antioxidant genes (p<0.001; Figure 1). The FC was 0.48 ± 0.07 for GPx, 0.53 ± 0.08 for GR, 0.45 ± 0.06 for GST, 0.58 ± 0.09 for GS, and 0.50 ± 0.07 for GCL in the IUGR samples (Figure 1).
- Glutathione-related enzyme gene expression in IUGR and HP groups. **p<0.0001 between FC values of each gene for the IUGR group compared to the HP group. FC, fold-change; HP, healthy pregnant; IUGR, intrauterine growth restriction.
Discussion
Superoxide anions, H2O2, and •OH are major ROS generated in electron transport chain reactions at the mitochondrial membrane.14,15 Increased lipid peroxidation rates, as shown by higher levels of MDA and H2O2, damage the cell membranes, and lead to a loss of cell integrity and function. The first step in ROS generation is SOA formation. Therefore, increased plasma or placental SOA levels presently noted in IUGR patients will result in OS, which may be a causative factor of IUGR.
Intrauterine growth restriction is considered a multifactorial disease potentially involving several risk factors, including social, cultural, environmental, and lifestyle factors. Social health determinants including low socioeconomic status, maternal age, education, inadequate prenatal care, pre-pregnancy weight age, short inter-pregnancy interval, smoking, and alcohol has considerable impact on IUGR. Such factors seem to vary with the economic status of the country. Approximately, 50% of the IUGR cases are due to reduced weight-gain, low BMI, if first time pregnant, short body build, and 25% cases are due to smoking habits in women in developed countries. Contrarily, cigarette smoking is considerably less in developing countries with an increased risk from other socio-health determinants.16 Other risk factors include infections, nutritional abnormalities, and chromosomal abnormalities, yet a significant number of cases are idiopathic.17 However, uteroplacental dysfunction due to decreased or a lack of maternal uteroplacental blood supply is a common leading cause of IUGR.18
Healthy placental functioning is essential for normal fetal development. Placental dysfunction restricts fetal growth. However, the exact pathophysiological processes underlying IUGR are still unclear. Approximately, 76% of intrauterine deaths are associated with IUGR.19 It is hypothesized that placental incompetency occurs early in pregnancy, at the time of trophoblast incorporation into the spiral arteries in the placental bed.20 This process requires increased energy to support cell division, development, and metabolism, which, in turn, generates excessive ROS, resulting in OS, and oxidative damage might play a crucial role in IUGR pathogenesis. In utero, the placenta is an important source of OS during pregnancy as polyunsaturated fatty acids are peroxidized into the maternal circulation. Oxidative stress is caused by an oxidant-antioxidant imbalance. Maternal consequences of IUGR could be due to free-radical generation because of defective spiral artery remodeling and decreased uteroplacental blood flow, as implicated in IUGR. Oxidative stress leads to placental insufficiency and subsequently to IUGR. To support this hypothesis, our results showed a significant increase in major oxidation markers in IUGR patients’ plasma and tissues samples and HP women. In addition, we found significantly decreased plasma and placental tissue activities, as well as significantly down-regulated expression levels of mRNA of major antioxidant glutathione-related enzymes in the placental tissue of IUGR patients compared to HP women. Therefore, the uncontrolled OS observed might have resulted in DNA damage, telomere shortening, and increased senescence of fetal membranes, leading to parturition, as previously described.15,21
During gestation, the antioxidant defense system quenches the free radicals generated in the placenta. Among major antioxidant enzymes, superoxide dismutase (SOD) encountered for primary defense mechanism, catalyzing the dismutation or reduction of SOA (O2•–) into H2O2. Hydrogen peroxide is then converted into water and oxygen by GPx, which eliminates H2O2 through the oxidization of GSH into GSSG. In addition to H2O2, GPx can oxidize GSH and reduces lipid and nonlipid hydroperoxides.5 In turn, GR, a flavor-protein enzyme, can use NADPH and result in GSH regeneration from GSSG, which allows the maintenance of a normal high intracellular GSH-GSSG ratio and preservation of the oxidative balance. The molar GSH-GSSG ratio is considered a cellular redox buffer indicating the cellular redox status22 and allows the maintenance of cellular homeostasis. When this balance changes in the direction of oxidative metabolism and excessive ROS generation, OS prevails, causing many diseases.23,24 Nicotinamide adenine dinucleotide phosphate and GR are the 2 most important components that regulate and maintain GSH levels (reduced-active form). The 39% decrease in the GSH-GSSG ratio in IUGR patients observed in this study could be due to decreased GSH and increased GSSG levels under OS.
Further, results demonstrated significant change in levels of glutathione related enzymes in IUGR patients compared to HP women. Plasma and placental GPx, GR, GST, GCL, and GS levels significantly decreased in IUGR patients’ (p<0.001), which is consistent with previous reports.25 Similarly, the enhanced lipid peroxidation and diminished antioxidant status observed are consistent with the results of recent studies.26 Hence, the literature and this study unanimously indicate lipid peroxide products as prominent risk factors of OS and IUGR.
The poor antioxidant status and diminished glutathione-related antioxidant enzyme activity observed in this study were confirmed by the recorded downregulation in the gene expression profiles of all studied enzymes. This downregulation could be due to the deleterious effect of ROS on trophoblastic cells, leading to gene inactivation with a net decrease in the transcription and protein synthesis of these enzymes. The mRNA profiling results were confirmed by H2O2, SOA, MDA, and PCC levels and GSH-GSSG ratio and decreased GSH levels in IUGR patients compared to HP women. All the OS markers studied indicated OS as a causative factor for IUGR.
The decrease observed in plasma and placental GSH levels in IUGR patients could be due to the significant decrease in GR activity or GSH’s reaction with the excessively generated H2O2, leading to increased GSSG formation. In addition, GSH levels decrease because of decreased rates of its GCL- and GS-catalyzed de novo synthesis, as previously described,27 both of which showed significantly decreased activity and significant downregulation of gene transcription.
In addition to the GSH-GSSG ratio, NAD+-NADH ratio is also essential for the cell redox state regulation and also within cell compartments, oxidative metabolism, mitochondrial function, gene transcripts, and signaling pathways.28,29 The decrease in NAD+ levels of IUGR patients might be due to excessive ROS generation, negatively affecting the NAD+ synthesis rate. This is coupled with increased NADH levels and a subsequent significant decrease in the NAD+-NADH ratio, in parallel with increased lipid peroxidation and PCC. These results mirror previous report24 who investigated changes in NAD metabolism in oxidatively stressed induced IUGR. However, one of current study limitations is that it did not include illustration of the exact causative mechanism, yet it strongly suggests the relevance of OS to IUGR conditions. To this end, a study to investigate the effects of excessive ROS levels and OS on the activities and gene transcripts of key NAD+ synthetic enzymes (nicotinamide/nicotinate mononucleotide adenyl-transferase; NMNAT, nicotinamide phosphoribosyl-transferase; NAMPT, and NAD+ synthase; NS), is underway in our laboratory. In addition, work is ongoing to examine IUGR placental activities and expression levels of sirtuin 1 as well as the activity of complex I-IV of the mitochondrial respiratory chain, which affect the redox state and the ATP production rate.
In conclusion, increased OS in placental tissue of IUGR patients decreases GSH-GSSG and NAD+-NADH ratios, decreases glutathione-related enzyme activity, and down-regulates their gene transcription. Although present work did not examine the underlying mechanism and the actual causative factors, the observed changes might play a crucial role in the IUGR etiology as the cellular redox state of plasma and placental tissue is adversely affected and shifts in favor of oxidative metabolism. Therefore, OS markers can be used as a diagnosis tool early in pregnancy or even in a planned conception for women with a history of IUGR. In addition, concentrated antioxidant supplementation treatment might decrease the risk for IUGR patients with a significant increase in OS markers; further studies on a large cohort to confirm this are required.
Acknowledgment
Authors extend their appreciation to the Deputyship for Research & Innovation, “Ministry of Education” in Saudi Arabia for funding this research work through the project number IFKSURG-1442-012. The authors also thank the Deanship of Scientific Research and RSSU at King Saud University for their technical support.
Footnotes
Disclosure. Authors extend their appreciation to the Deputyship for Research & Innovation, “Ministry of Education” in Saudi Arabia for funding this research work through the project number IFKSURG-1442-012. The authors also thank the Deanship of Scientific Research and RSSU at King Saud University for their technical support.
- Received October 27, 2020.
- Accepted March 15, 2021.
- Copyright: © Saudi Medical Journal
This is an open-access article distributed under the terms of the Creative Commons Attribution-Noncommercial License (CC BY-NC), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.